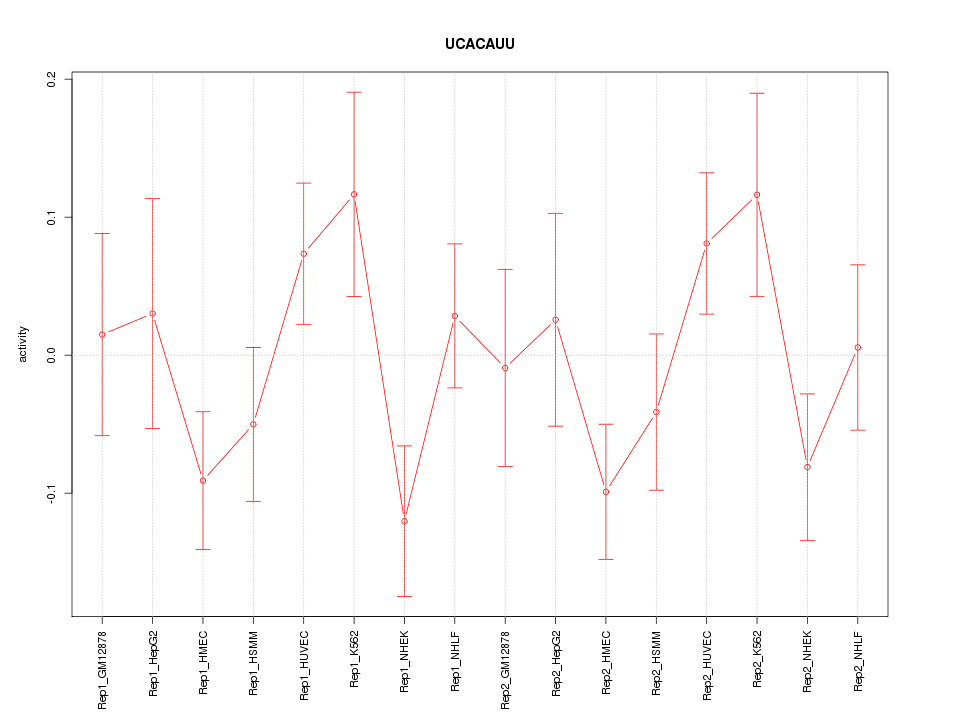
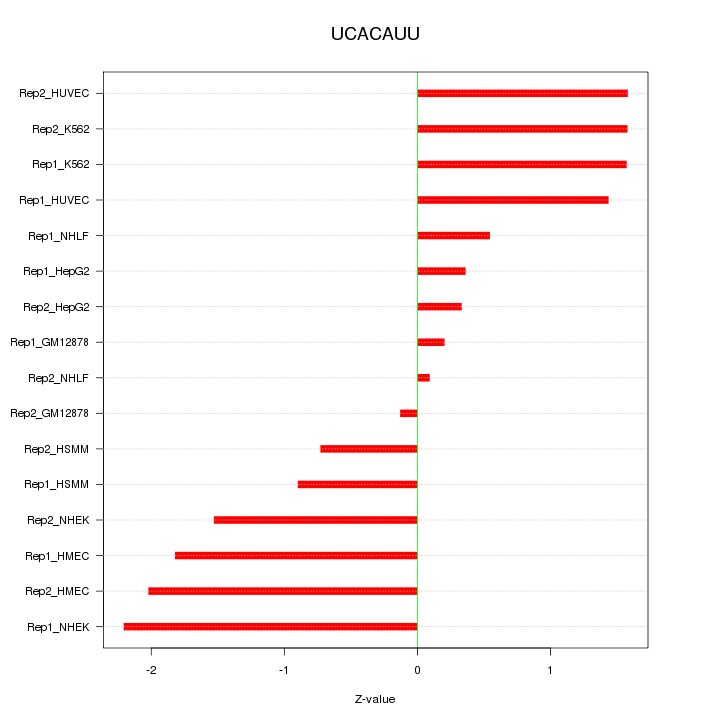
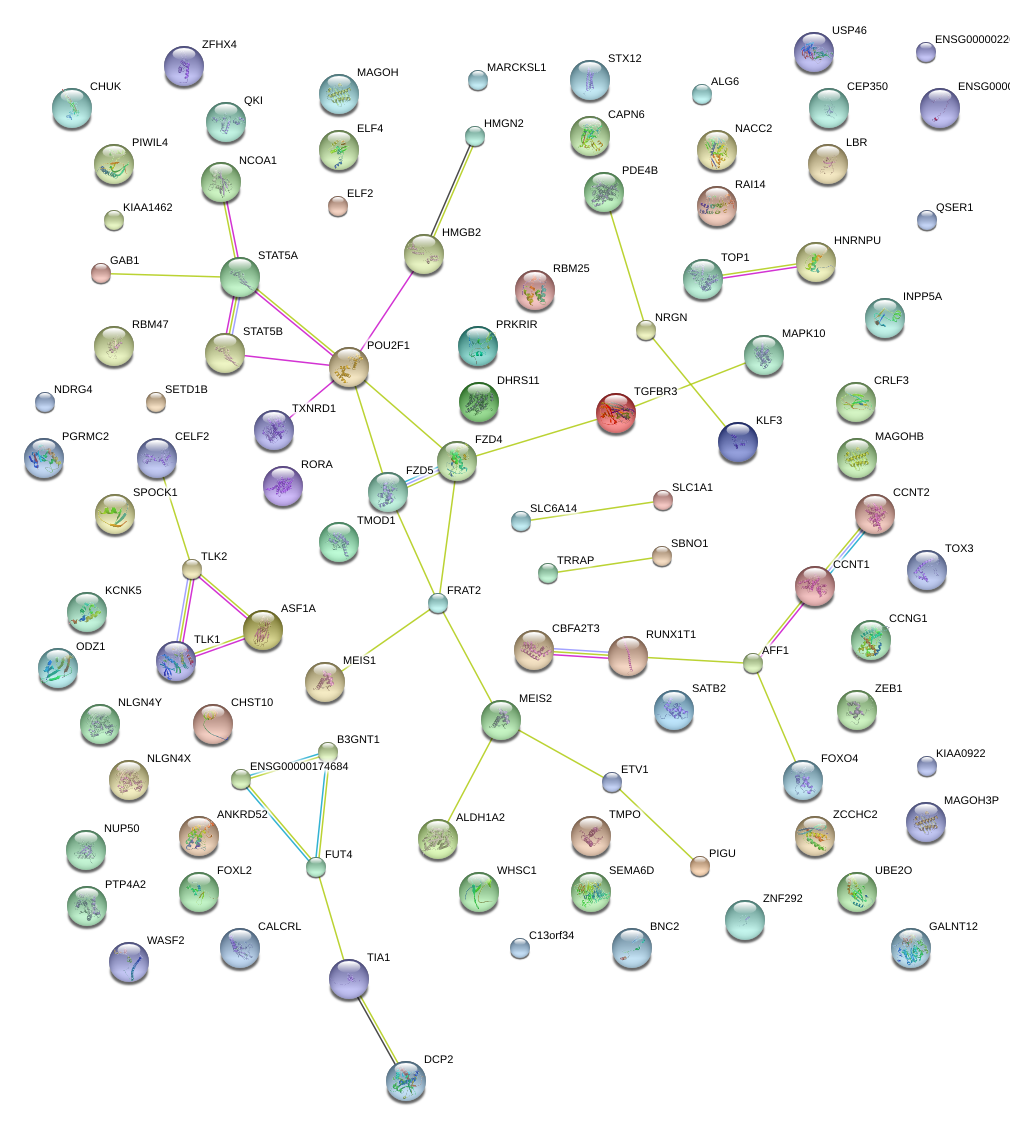
|
chr1_-_92351613
|
4.356
|
NM_001195683
NM_003243
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_92371558
|
3.652
|
NM_001195684
|
TGFBR3
|
transforming growth factor, beta receptor III
|
|
chr1_-_32801606
|
2.073
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr16_-_89043215
|
1.779
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr16_-_89007605
|
1.663
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr4_-_174254730
|
1.519
|
NM_001130689
|
HMGB2
|
high mobility group box 2
|
|
chr3_-_138665981
|
1.452
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr4_-_174255454
|
1.304
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr2_-_101034049
|
1.239
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr17_-_40428391
|
1.207
|
NM_012448
|
STAT5B
|
signal transducer and activator of transcription 5B
|
|
chr2_-_188312872
|
1.146
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr9_-_138987096
|
1.140
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr9_+_101569933
|
1.125
|
NM_024642
|
GALNT12
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12)
|
|
chr2_-_200329810
|
1.051
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr2_-_200322699
|
1.021
|
NM_001172509
|
SATB2
|
SATB homeobox 2
|
|
chr6_-_39197225
|
1.008
|
NM_003740
|
KCNK5
|
potassium channel, subfamily K, member 5
|
|
chr1_+_66797775
|
1.008
|
NM_001037339
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr6_+_119215240
|
0.959
|
NM_014034
|
ASF1A
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr1_+_66458389
|
0.948
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chrY_+_16634487
|
0.933
|
NM_001206850
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr1_+_66258192
|
0.922
|
NM_001037341
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr2_-_200335988
|
0.901
|
NM_015265
|
SATB2
|
SATB homeobox 2
|
|
chr1_-_32403975
|
0.896
|
NM_080391
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr17_+_34948213
|
0.865
|
NM_024308
|
DHRS11
|
dehydrogenase/reductase (SDR family) member 11
|
|
chr7_+_98476084
|
0.863
|
NM_001244580
NM_003496
|
TRRAP
|
transformation/transcription domain-associated protein
|
|
chr1_+_66258855
|
0.852
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr5_+_112312392
|
0.813
|
NM_001242377
NM_152624
|
DCP2
|
DCP2 decapping enzyme homolog (S. cerevisiae)
|
|
chrY_+_16635625
|
0.707
|
NM_014893
|
NLGN4Y
|
neuroligin 4, Y-linked
|
|
chr4_+_38665587
|
0.699
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr11_-_86666211
|
0.695
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr4_-_140060601
|
0.686
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr11_-_66115016
|
0.672
|
NM_006876
|
B3GNT1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr8_-_93111915
|
0.668
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_32385258
|
0.664
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr22_+_45560529
|
0.648
|
NM_153645
|
NUP50
|
nucleoporin 50kDa
|
|
chr15_-_58358120
|
0.637
|
NM_001206897
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr12_+_104680712
|
0.616
|
NM_003330
NM_182729
NM_182742
NM_182743
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr4_+_154387450
|
0.612
|
NM_001131007
NM_015196
|
KIAA0922
|
KIAA0922
|
|
chr1_-_245027808
|
0.598
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr12_-_123834984
|
0.587
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr2_-_208634051
|
0.585
|
NM_003468
|
FZD5
|
frizzled family receptor 5
|
|
chr1_+_63833260
|
0.575
|
NM_013339
|
ALG6
|
asparagine-linked glycosylation 6, alpha-1,3-glucosyltransferase homolog (S. cerevisiae)
|
|
chr1_+_179923870
|
0.567
|
NM_014810
|
CEP350
|
centrosomal protein 350kDa
|
|
chr5_+_162864571
|
0.566
|
NM_004060
NM_199246
|
CCNG1
|
cyclin G1
|
|
chr17_-_74449272
|
0.550
|
NM_022066
|
UBE2O
|
ubiquitin-conjugating enzyme E2O
|
|
chr18_+_60190657
|
0.548
|
NM_017742
|
ZCCHC2
|
zinc finger, CCHC domain containing 2
|
|
chr8_-_93029864
|
0.537
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_-_93107881
|
0.530
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr12_-_10766130
|
0.517
|
NM_018048
|
MAGOHB
|
mago-nashi homolog B (Drosophila)
|
|
chr10_+_11047253
|
0.513
|
NM_001025077
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr15_-_58306185
|
0.496
|
NM_170697
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr1_+_28099682
|
0.487
|
NM_177424
|
STX12
|
syntaxin 12
|
|
chr6_+_87865261
|
0.483
|
NM_015021
|
ZNF292
|
zinc finger protein 292
|
|
chrX_-_124097601
|
0.473
|
NM_001163278
NM_001163279
NM_014253
|
ODZ1
|
odz, odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr12_+_122242580
|
0.471
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|
|
chr12_+_104609501
|
0.470
|
NM_001093771
|
TXNRD1
|
thioredoxin reductase 1
|
|
chr10_+_134351272
|
0.467
|
NM_005539
|
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa
|
|
chr11_-_76091866
|
0.465
|
NM_004705
|
PRKRIR
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor)
|
|
chr10_-_30348433
|
0.465
|
NM_020848
|
KIAA1462
|
KIAA1462
|
|
chr13_+_73301980
|
0.464
|
NM_024808
|
BORA
|
bora, aurora kinase A activator
|
|
chr8_-_93075190
|
0.450
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr8_+_77593506
|
0.450
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr1_-_27816676
|
0.442
|
NM_001201404
NM_006990
|
WASF2
|
WAS protein family, member 2
|
|
chr9_+_4490193
|
0.441
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr9_-_16870719
|
0.427
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr8_-_93115453
|
0.417
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr16_-_52580805
|
0.409
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr8_-_93107442
|
0.409
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_-_140005464
|
0.408
|
NM_006874
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr15_-_60884615
|
0.404
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_60919642
|
0.402
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr12_-_56652118
|
0.398
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chrX_-_110513750
|
0.396
|
NM_014289
|
CAPN6
|
calpain 6
|
|
chr5_+_34684611
|
0.394
|
NM_001145521
NM_001145525
|
RAI14
|
retinoic acid induced 14
|
|
chr2_+_66662516
|
0.394
|
NM_002398
|
MEIS1
|
Meis homeobox 1
|
|
chr11_+_124609775
|
0.394
|
NM_001126181
NM_006176
|
NRGN
|
neurogranin (protein kinase C substrate, RC3)
|
|
chr11_+_32914359
|
0.393
|
NM_001076786
|
QSER1
|
glutamine and serine rich 1
|
|
chr15_-_61521478
|
0.390
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_+_34656595
|
0.390
|
NM_001145520
|
RAI14
|
retinoic acid induced 14
|
|
chr10_-_99094427
|
0.388
|
NM_012083
|
FRAT2
|
frequently rearranged in advanced T-cell lymphomas 2
|
|
chr16_-_52581713
|
0.380
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr10_+_11059854
|
0.380
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chrX_-_129244471
|
0.380
|
NM_001421
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr2_+_135676387
|
0.375
|
NM_001241
NM_058241
|
CCNT2
|
cyclin T2
|
|
chr4_+_144257982
|
0.374
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr4_+_87856152
|
0.372
|
NM_001166693
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr4_+_87928083
|
0.370
|
NM_005935
|
AFF1
|
AF4/FMR2 family, member 1
|
|
chr4_-_129209983
|
0.364
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr2_-_70475563
|
0.363
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr1_-_225616556
|
0.360
|
NM_194442
|
LBR
|
lamin B receptor
|
|
chr1_-_225615787
|
0.359
|
NM_002296
|
LBR
|
lamin B receptor
|
|
chr10_-_101989342
|
0.358
|
NM_001278
|
CHUK
|
conserved helix-loop-helix ubiquitous kinase
|
|
chr14_+_73525187
|
0.351
|
NM_021239
|
RBM25
|
RNA binding motif protein 25
|
|
chr9_+_100263818
|
0.343
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr2_-_172087823
|
0.343
|
NM_001136554
|
TLK1
|
tousled-like kinase 1
|
|
chr1_+_167298280
|
0.342
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_+_163835668
|
0.340
|
NM_006775
NM_206853
NM_206854
NM_206855
|
QKI
|
QKI, KH domain containing, RNA binding
|
|
chr15_+_48010685
|
0.340
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr4_-_53525316
|
0.336
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr4_-_40631846
|
0.335
|
NM_001098634
|
RBM47
|
RNA binding motif protein 47
|
|
chr22_+_45559725
|
0.327
|
NM_007172
|
NUP50
|
nucleoporin 50kDa
|
|
chr12_+_98909331
|
0.325
|
NM_001032283
NM_001032284
NM_003276
|
TMPO
|
thymopoietin
|
|
chr16_+_58534045
|
0.324
|
NM_001242833
NM_001242834
NM_001242835
NM_001242836
|
NDRG4
|
NDRG family member 4
|
|
chr2_+_24807345
|
0.324
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr2_-_171923482
|
0.321
|
NM_001136555
|
TLK1
|
tousled-like kinase 1
|
|
chr10_+_31610063
|
0.320
|
NM_001128128
NM_001174094
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chr10_+_31608097
|
0.317
|
NM_001174093
NM_001174095
NM_001174096
NM_030751
|
ZEB1
|
zinc finger E-box binding homeobox 1
|
|
chrX_-_129244652
|
0.311
|
NM_001127197
|
ELF4
|
E74-like factor 4 (ets domain transcription factor)
|
|
chr5_+_34656367
|
0.301
|
NM_001145522
NM_015577
|
RAI14
|
retinoic acid induced 14
|
|
chr7_-_14025826
|
0.299
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chrX_+_70315637
|
0.299
|
NM_001170931
NM_005938
|
FOXO4
|
forkhead box O4
|
|
chr11_+_94277005
|
0.296
|
NM_002033
|
PIWIL4
FUT4
|
piwi-like 4 (Drosophila)
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific)
|
|
chrX_+_115567746
|
0.295
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr1_+_26798901
|
0.294
|
NM_005517
|
HMGN2
|
high mobility group nucleosomal binding domain 2
|
|
chr20_+_39657461
|
0.294
|
NM_003286
|
TOP1
|
topoisomerase (DNA) I
|
|
chr17_-_29151724
|
0.291
|
NM_015986
|
CRLF3
|
cytokine receptor-like factor 3
|
|
chr9_+_100263461
|
0.289
|
NM_001166116
|
TMOD1
|
tropomodulin 1
|
|
chr5_-_136834887
|
0.289
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr4_+_1894508
|
0.279
|
NM_007331
NM_133330
NM_133331
NM_133335
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr5_-_88199868
|
0.276
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr6_-_24719286
|
0.274
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr16_-_4897265
|
0.273
|
NM_032569
|
GLYR1
|
glyoxylate reductase 1 homolog (Arabidopsis)
|
|
chr10_+_81107219
|
0.273
|
NM_005729
|
PPIF
|
peptidylprolyl isomerase F
|
|
chr5_+_34687663
|
0.272
|
NM_001145523
|
RAI14
|
retinoic acid induced 14
|
|
chr5_-_88179278
|
0.268
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_186064416
|
0.266
|
NM_001151
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4
|
|
chr1_+_23037329
|
0.265
|
NM_004442
NM_017449
|
EPHB2
|
EPH receptor B2
|
|
chr10_-_101492422
|
0.265
|
NM_004376
NM_078470
|
COX15
|
COX15 homolog, cytochrome c oxidase assembly protein (yeast)
|
|
chr2_-_47142950
|
0.262
|
NM_001171506
NM_001171509
NM_139279
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr20_-_23066828
|
0.261
|
NM_012072
|
CD93
|
CD93 molecule
|
|
chr10_+_89623091
|
0.260
|
NM_000314
|
PTEN
|
phosphatase and tensin homolog
|
|
chr18_+_9913954
|
0.259
|
NM_003574
NM_194434
|
VAPA
|
VAMP (vesicle-associated membrane protein)-associated protein A, 33kDa
|
|
chr6_-_35696359
|
0.257
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr16_+_58497548
|
0.257
|
NM_020465
|
NDRG4
|
NDRG family member 4
|
|
chr2_-_47143250
|
0.256
|
NM_001171507
NM_001171510
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr2_+_88991166
|
0.255
|
NM_144563
|
RPIA
|
ribose 5-phosphate isomerase A
|
|
chr17_-_10372875
|
0.251
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr16_+_15068832
|
0.251
|
NM_015027
|
PDXDC1
|
pyridoxal-dependent decarboxylase domain containing 1
|
|
chr12_+_32654976
|
0.250
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr1_+_167189948
|
0.245
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr9_-_94124194
|
0.244
|
NM_001698
|
AUH
|
AU RNA binding protein/enoyl-CoA hydratase
|
|
chr6_+_151646634
|
0.242
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr4_+_140222668
|
0.237
|
NM_057175
|
NAA15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr1_+_65730423
|
0.237
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr17_-_65241305
|
0.236
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr16_+_58497993
|
0.236
|
NM_001130487
|
NDRG4
|
NDRG family member 4
|
|
chr11_+_86748870
|
0.236
|
NM_001168724
NM_022918
|
TMEM135
|
transmembrane protein 135
|
|
chr7_-_14029641
|
0.232
|
NM_001163147
NM_004956
|
ETV1
|
ets variant 1
|
|
chr10_-_65028825
|
0.231
|
NM_004241
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr2_-_225450105
|
0.229
|
NM_003590
|
CUL3
|
cullin 3
|
|
chrX_+_122993536
|
0.227
|
NM_001204401
NM_001167
|
XIAP
|
X-linked inhibitor of apoptosis
|
|
chr17_+_58677489
|
0.222
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr7_-_139876718
|
0.222
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr15_-_52587851
|
0.221
|
NM_018728
|
MYO5C
|
myosin VC
|
|
chr11_-_47545539
|
0.219
|
NM_001172639
|
CELF1
|
CUGBP, Elav-like family member 1
|
|
chr1_-_247335251
|
0.213
|
NM_001243740
NM_003431
|
ZNF124
|
zinc finger protein 124
|
|
chr16_-_58231641
|
0.212
|
NM_001896
|
CSNK2A2
|
casein kinase 2, alpha prime polypeptide
|
|
chr2_-_47168902
|
0.209
|
NM_001171508
NM_001171511
|
MCFD2
|
multiple coagulation factor deficiency 2
|
|
chr20_+_37434294
|
0.207
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory subunit 16B
|
|
chr12_+_72233486
|
0.206
|
NM_001146213
NM_001146214
NM_022771
|
TBC1D15
|
TBC1 domain family, member 15
|
|
chr3_-_50365663
|
0.205
|
NM_007275
|
TUSC2
|
tumor suppressor candidate 2
|
|
chr10_+_30722856
|
0.202
|
NM_001244134
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr8_-_60031545
|
0.202
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr1_-_231560789
|
0.200
|
NM_022051
|
EGLN1
|
egl nine homolog 1 (C. elegans)
|
|
chr14_+_103058988
|
0.200
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr10_+_11206928
|
0.198
|
NM_001025076
NM_001083591
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr5_+_127419407
|
0.198
|
NM_001046
|
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporters), member 2
|
|
chr5_+_72143929
|
0.198
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chr1_-_51984994
|
0.196
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr13_+_20532788
|
0.195
|
NM_003453
NM_197968
NM_001190964
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr4_+_1873035
|
0.195
|
NM_001042424
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr7_+_117824085
|
0.194
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr13_+_20532906
|
0.192
|
NM_001190965
|
ZMYM2
|
zinc finger, MYM-type 2
|
|
chr18_-_55253785
|
0.192
|
NM_000140
NM_001012515
|
FECH
|
ferrochelatase
|
|
chr10_-_65225605
|
0.191
|
NM_032776
|
JMJD1C
|
jumonji domain containing 1C
|
|
chr1_+_24286300
|
0.188
|
NM_017761
|
PNRC2
|
proline-rich nuclear receptor coactivator 2
|
|
chr5_+_56110899
|
0.188
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr1_-_150669499
|
0.187
|
NM_018178
|
GOLPH3L
|
golgi phosphoprotein 3-like
|
|
chr19_-_48744276
|
0.186
|
NM_001184900
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr17_-_38574042
|
0.183
|
NM_001067
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa
|
|
chr19_-_48752921
|
0.182
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr1_-_55681024
|
0.178
|
NM_015306
|
USP24
|
ubiquitin specific peptidase 24
|
|
chr2_-_21022812
|
0.172
|
NM_021925
|
C2orf43
|
chromosome 2 open reading frame 43
|
|
chr2_-_172017358
|
0.168
|
NM_012290
|
TLK1
|
tousled-like kinase 1
|
|
chr12_+_81471796
|
0.166
|
NM_024560
|
ACSS3
|
acyl-CoA synthetase short-chain family member 3
|
|
chr4_-_76439582
|
0.166
|
NM_001008925
NM_001009922
NM_015436
|
RCHY1
|
ring finger and CHY zinc finger domain containing 1
|
|
chrX_+_64708614
|
0.166
|
NM_001010888
|
ZC3H12B
|
zinc finger CCCH-type containing 12B
|
|
chr3_-_66550844
|
0.165
|
NM_015541
|
LRIG1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr8_-_8750739
|
0.165
|
NM_004225
|
MFHAS1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr8_+_61591239
|
0.164
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr11_+_75526211
|
0.164
|
NM_003369
|
UVRAG
|
UV radiation resistance associated gene
|
|
chr1_-_68962726
|
0.162
|
NM_001114120
NM_017779
|
DEPDC1
|
DEP domain containing 1
|
|
chr2_-_37193609
|
0.161
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr2_+_152214105
|
0.161
|
NM_007115
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6
|
|
chr3_+_38495778
|
0.160
|
NM_001106
|
ACVR2B
|
activin A receptor, type IIB
|
|
chr12_+_74931550
|
0.160
|
NM_001136262
|
ATXN7L3B
|
ataxin 7-like 3B
|
|
chr10_+_99079017
|
0.158
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr1_+_159141376
|
0.157
|
NM_021189
NM_001127173
|
CADM3
|
cell adhesion molecule 3
|
|
chr16_+_31885077
|
0.157
|
NM_003414
|
ZNF267
|
zinc finger protein 267
|
|
chr12_+_12938540
|
0.157
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|