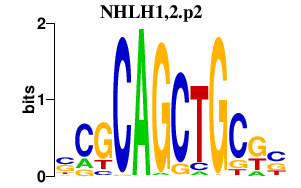
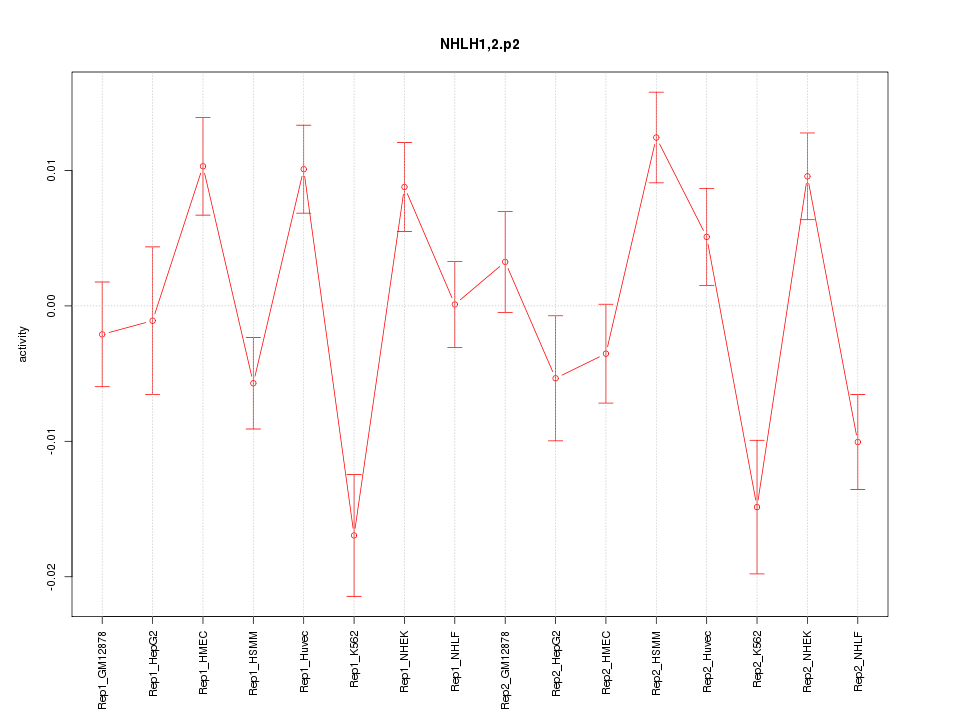
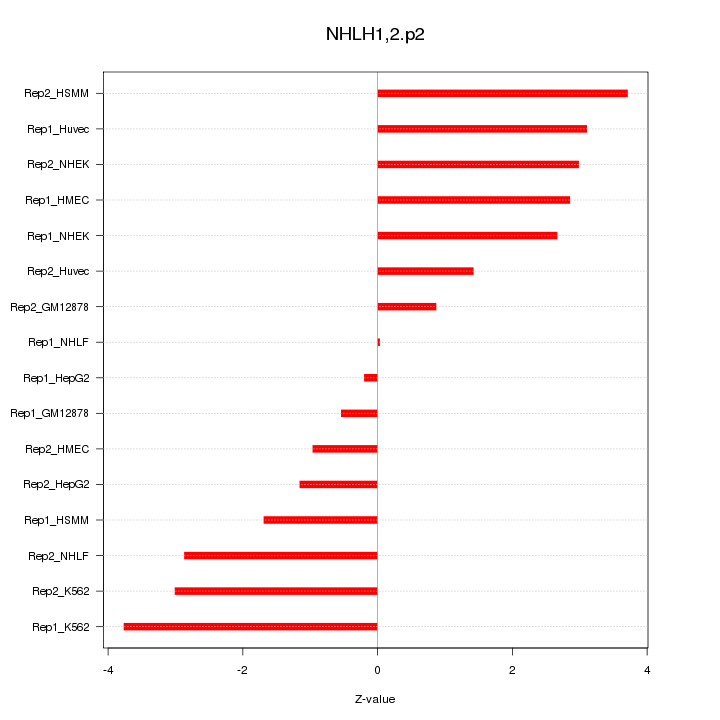
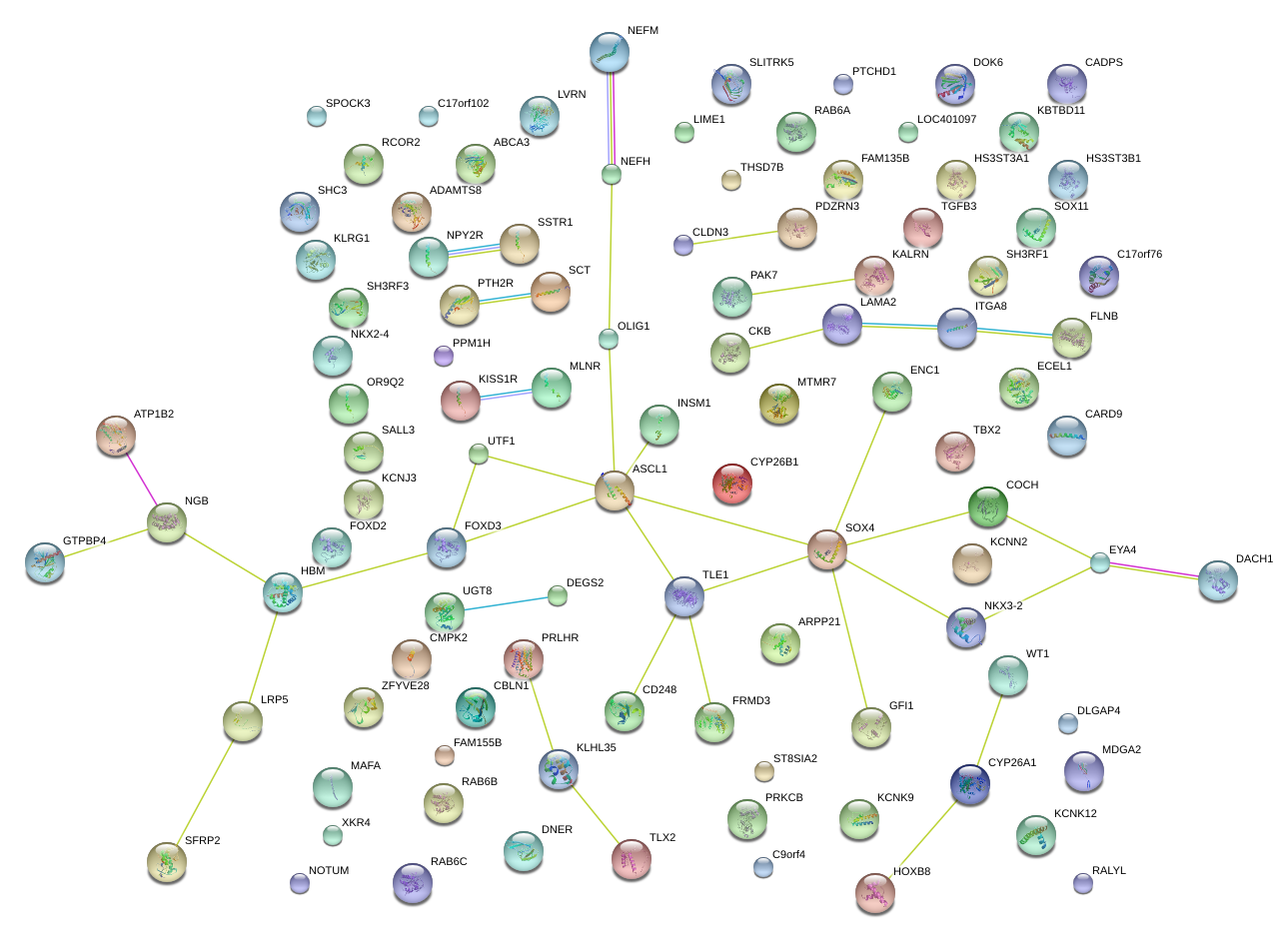
|
chr8_+_1937711
|
3.333
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr16_+_155972
|
3.311
|
NM_001003938
|
HBM
|
hemoglobin, mu
|
|
chr8_-_24828134
|
3.249
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr5_+_113726203
|
2.878
|
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr5_+_113725914
|
2.715
|
NM_021614
|
KCNN2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr14_-_103058568
|
2.576
|
|
CKB
|
creatine kinase, brain
|
|
chr2_-_6923215
|
2.505
|
NM_207315
|
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial
|
|
chr3_-_62835601
|
2.490
|
|
|
|
|
chr13_-_71338144
|
2.396
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr7_-_72822471
|
2.291
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr13_+_48692472
|
2.250
|
NM_001507
|
MLNR
|
motilin receptor
|
|
chr2_+_74595073
|
2.172
|
NM_016170
|
TLX2
|
T-cell leukemia homeobox 2
|
|
chr20_+_20296744
|
2.090
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr4_-_13155135
|
2.077
|
NM_001189
|
NKX3-2
|
NK3 homeobox 2
|
|
chr21_+_33364420
|
2.014
|
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chrX_+_23262905
|
2.008
|
NM_173495
|
PTCHD1
|
patched domain containing 1
|
|
chr14_+_30413393
|
1.909
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr17_+_56832492
|
1.900
|
|
TBX2
|
T-box 2
|
|
chr17_-_77512346
|
1.852
|
NM_178493
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila)
|
|
chr8_+_85258007
|
1.823
|
NM_001100392
NM_001100393
|
RALYL
|
RALY RNA binding protein-like
|
|
chr10_-_15801661
|
1.815
|
NM_003638
|
ITGA8
|
integrin, alpha 8
|
|
chr8_+_56177567
|
1.807
|
NM_052898
|
XKR4
|
XK, Kell blood group complex subunit-related family, member 4
|
|
chr2_+_208979800
|
1.805
|
NM_005048
|
PTH2R
|
parathyroid hormone 2 receptor
|
|
chr3_-_73756668
|
1.793
|
NM_015009
|
PDZRN3
|
PDZ domain containing ring finger 3
|
|
chr17_+_7494964
|
1.779
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr11_-_63440891
|
1.764
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chr8_-_144862266
|
1.754
|
NM_001162914
|
LOC100130274
|
coiled-coil domain containing 121-like
|
|
chr6_+_133604203
|
1.682
|
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr10_+_94823636
|
1.663
|
NM_000783
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr3_+_161426322
|
1.643
|
|
LOC401097
|
hypothetical protein LOC401097
|
|
chr2_+_109112428
|
1.638
|
NM_001099289
|
SH3RF3
|
SH3 domain containing ring finger 3
|
|
chr8_+_85258056
|
1.629
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr16_+_23754860
|
1.607
|
|
PRKCB
|
protein kinase C, beta
|
|
chr15_+_90738108
|
1.597
|
NM_006011
|
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2
|
|
chr1_-_92724181
|
1.588
|
NM_001127216
|
GFI1
|
growth factor independent 1 transcription repressor
|
|
chr11_-_74818685
|
1.584
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr22_-_37569679
|
1.568
|
|
|
|
|
chr8_+_24827181
|
1.554
|
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr13_-_71338558
|
1.535
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr17_-_16335809
|
1.512
|
|
C17orf76
|
chromosome 17 open reading frame 76
|
|
chr3_-_62836052
|
1.495
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chr10_+_134893766
|
1.483
|
NM_003577
|
UTF1
|
undifferentiated embryonic cell transcription factor 1
|
|
chr14_-_99695684
|
1.475
|
NM_206918
|
DEGS2
|
degenerative spermatocyte homolog 2, lipid desaturase (Drosophila)
|
|
chr2_-_47650973
|
1.473
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr19_+_868285
|
1.471
|
NM_032551
|
KISS1R
|
KISS1 receptor
|
|
chr8_+_24827173
|
1.469
|
NM_005382
|
NEFM
|
neurofilament, medium polypeptide
|
|
chr18_+_74841262
|
1.456
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chrX_+_68641802
|
1.438
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr14_+_37748345
|
1.428
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr2_-_72228427
|
1.426
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr9_-_83493415
|
1.408
|
NM_005077
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila)
|
|
chr14_-_76807302
|
1.407
|
NM_021257
|
NGB
|
neuroglobin
|
|
chr4_-_168392307
|
1.399
|
NM_001040159
NM_016950
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr16_+_23754997
|
1.398
|
|
PRKCB
|
protein kinase C, beta
|
|
chr2_-_233060774
|
1.396
|
NM_004826
|
ECEL1
|
endothelin converting enzyme-like 1
|
|
chr6_+_129245978
|
1.394
|
NM_000426
NM_001079823
|
LAMA2
|
laminin, alpha 2
|
|
chr11_-_74818858
|
1.393
|
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr9_-_110969310
|
1.390
|
NM_014334
|
C9orf4
|
chromosome 9 open reading frame 4
|
|
chr11_-_129803515
|
1.381
|
|
ADAMTS8
|
ADAM metallopeptidase with thrombospondin type 1 motif, 8
|
|
chr3_+_35656012
|
1.372
|
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr2_+_137239583
|
1.363
|
|
THSD7B
|
thrombospondin, type I, domain containing 7B
|
|
chr17_-_44047299
|
1.357
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr20_-_21326046
|
1.346
|
NM_033176
|
NKX2-4
|
NK2 homeobox 4
|
|
chr13_+_87122778
|
1.338
|
NM_015567
|
SLITRK5
|
SLIT and NTRK-like family, member 5
|
|
chr2_-_230287442
|
1.335
|
NM_139072
|
DNER
|
delta/notch-like EGF repeat containing
|
|
chr18_+_65219114
|
1.334
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr11_-_617172
|
1.327
|
NM_021920
|
SCT
|
secretin
|
|
chr5_+_115326035
|
1.317
|
NM_173800
|
AQPEP
|
laeverin
|
|
chr4_+_115739006
|
1.311
|
NM_001128174
|
UGT8
|
UDP glycosyltransferase 8
|
|
chr4_-_168392266
|
1.308
|
|
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3
|
|
chr20_-_9767406
|
1.298
|
NM_020341
NM_177990
|
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr22_+_28206180
|
1.298
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr17_+_56832255
|
1.285
|
|
TBX2
|
T-box 2
|
|
chr14_-_47213906
|
1.269
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr14_+_37748031
|
1.264
|
|
SSTR1
|
somatostatin receptor 1
|
|
chr16_-_47872880
|
1.263
|
|
|
|
|
chr11_-_65841077
|
1.257
|
NM_020404
|
CD248
|
CD248 molecule, endosialin
|
|
chr12_-_61615124
|
1.253
|
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr4_-_2390050
|
1.237
|
NM_001172656
NM_001172657
NM_001172658
NM_020972
|
ZFYVE28
|
zinc finger, FYVE domain containing 28
|
|
chr12_+_101876151
|
1.231
|
|
ASCL1
|
achaete-scute complex homolog 1 (Drosophila)
|
|
chr17_-_16336204
|
1.229
|
NM_001113567
NM_207387
|
C17orf76
|
chromosome 17 open reading frame 76
|
|
chr11_-_32413486
|
1.224
|
|
WT1
|
Wilms tumor 1
|
|
chr11_-_32413635
|
1.223
|
NM_000378
NM_024424
NM_024426
|
WT1
|
Wilms tumor 1
|
|
chr2_+_155263338
|
1.217
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr17_+_14145051
|
1.216
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr9_-_90983195
|
1.213
|
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr8_-_144583718
|
1.212
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr9_-_85342868
|
1.197
|
NM_174938
|
FRMD3
|
FERM domain containing 3
|
|
chr16_-_47873183
|
1.195
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr3_+_125786195
|
1.186
|
NM_007064
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr4_+_156349230
|
1.186
|
NM_000910
|
NPY2R
|
neuropeptide Y receptor Y2
|
|
chr20_+_34357604
|
1.178
|
|
DLGAP4
|
discs, large (Drosophila) homolog-associated protein 4
|
|
chr4_-_154929657
|
1.155
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr17_-_29930431
|
1.152
|
NM_207454
|
C17orf102
|
chromosome 17 open reading frame 102
|
|
chr11_-_74819165
|
1.147
|
NM_001039548
|
KLHL35
|
kelch-like 35 (Drosophila)
|
|
chr8_-_17315386
|
1.146
|
NM_004686
|
MTMR7
|
myotubularin related protein 7
|
|
chr16_+_23754772
|
1.143
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr2_+_5750229
|
1.142
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr8_-_139578246
|
1.141
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr8_-_140784416
|
1.139
|
NM_016601
|
KCNK9
|
potassium channel, subfamily K, member 9
|
|
chr1_+_63561632
|
1.132
|
|
FOXD3
|
forkhead box D3
|
|
chr3_-_135097111
|
1.132
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr16_+_23754822
|
1.127
|
|
PRKCB
|
protein kinase C, beta
|
|
chr10_-_120345148
|
1.119
|
NM_004248
|
PRLHR
|
prolactin releasing hormone receptor
|
|
chr4_+_61749540
|
1.115
|
|
LPHN3
|
latrophilin 3
|
|
chr3_-_10724715
|
1.113
|
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr7_+_98084531
|
1.112
|
NM_002523
|
NPTX2
|
neuronal pentraxin II
|
|
chr1_-_21868380
|
1.110
|
NM_001145657
NM_002885
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chr9_-_139183675
|
1.098
|
|
|
|
|
chrX_+_117992623
|
1.090
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chrX_-_99551926
|
1.087
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr1_+_1062137
|
1.077
|
|
LOC254099
|
hypothetical protein LOC254099
|
|
chr19_-_51856129
|
1.073
|
NM_145056
|
DACT3
|
dapper, antagonist of beta-catenin, homolog 3 (Xenopus laevis)
|
|
chr11_+_13940715
|
1.073
|
|
SPON1
|
spondin 1, extracellular matrix protein
|
|
chr3_-_135096986
|
1.062
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr19_-_6453224
|
1.059
|
NM_006087
|
TUBB4
|
tubulin, beta 4
|
|
chr8_+_31616809
|
1.055
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr6_+_124166767
|
1.052
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr12_+_131705438
|
1.046
|
NM_012226
NM_016318
NM_170682
NM_170683
NM_174872
NM_174873
|
P2RX2
|
purinergic receptor P2X, ligand-gated ion channel, 2
|
|
chr3_-_124650081
|
1.045
|
NM_183357
|
ADCY5
|
adenylate cyclase 5
|
|
chr16_-_47872973
|
1.041
|
|
CBLN1
|
cerebellin 1 precursor
|
|
chr16_+_270606
|
1.038
|
NM_001176
|
ARHGDIG
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr22_+_28446288
|
1.036
|
NM_182527
|
CABP7
|
calcium binding protein 7
|
|
chr4_-_155631861
|
1.034
|
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr17_-_17340219
|
1.033
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr12_+_5023345
|
1.032
|
NM_002234
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5
|
|
chr8_+_35212505
|
1.029
|
NM_080872
|
UNC5D
|
unc-5 homolog D (C. elegans)
|
|
chr3_-_30911156
|
1.026
|
NM_207359
|
GADL1
|
glutamate decarboxylase-like 1
|
|
chr20_+_13150417
|
1.025
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr1_+_229365296
|
1.019
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr21_+_18539016
|
1.018
|
NM_024944
|
CHODL
|
chondrolectin
|
|
chr17_-_7106497
|
1.012
|
|
CLDN7
|
claudin 7
|
|
chr22_+_16973399
|
1.009
|
NM_001193414
NM_018943
|
TUBA8
|
tubulin, alpha 8
|
|
chr22_+_42138034
|
1.002
|
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr19_-_14177980
|
0.995
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr14_-_37795290
|
0.993
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr14_-_60185659
|
0.986
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr10_+_23521464
|
0.984
|
NM_178161
|
PTF1A
|
pancreas specific transcription factor, 1a
|
|
chr2_-_128792515
|
0.983
|
NM_004807
|
HS6ST1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chrX_+_21302317
|
0.983
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr5_+_80292313
|
0.983
|
NM_006909
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr3_-_69064364
|
0.981
|
NM_001005527
NM_182522
|
FAM19A4
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A4
|
|
chr10_-_99521701
|
0.980
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr7_-_30688626
|
0.970
|
NM_001883
|
CRHR2
|
corticotropin releasing hormone receptor 2
|
|
chr4_+_177223978
|
0.968
|
NM_170710
NM_181265
|
WDR17
|
WD repeat domain 17
|
|
chr9_+_94987032
|
0.967
|
NM_006648
|
WNK2
|
WNK lysine deficient protein kinase 2
|
|
chr2_+_185171480
|
0.965
|
|
ZNF804A
|
zinc finger protein 804A
|
|
chr11_+_14621972
|
0.964
|
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr7_-_45927263
|
0.955
|
NM_000598
NM_001013398
|
IGFBP3
|
insulin-like growth factor binding protein 3
|
|
chr8_-_17315171
|
0.949
|
|
MTMR7
|
myotubularin related protein 7
|
|
chr10_+_105026773
|
0.947
|
NM_032727
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr19_-_44386731
|
0.945
|
NM_001080468
|
SYCN
|
syncollin
|
|
chr10_-_135000358
|
0.941
|
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr20_+_19141364
|
0.937
|
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr4_-_85638410
|
0.929
|
NM_006168
|
NKX6-1
|
NK6 homeobox 1
|
|
chr5_+_170779664
|
0.927
|
|
FGF18
|
fibroblast growth factor 18
|
|
chr17_+_29931880
|
0.918
|
NM_207313
|
TMEM132E
|
transmembrane protein 132E
|
|
chrX_+_12066505
|
0.914
|
NM_014728
|
FRMPD4
|
FERM and PDZ domain containing 4
|
|
chr20_-_49852353
|
0.905
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr22_-_29231597
|
0.904
|
NM_001161368
NM_174977
|
SEC14L4
|
SEC14-like 4 (S. cerevisiae)
|
|
chr4_+_3738093
|
0.904
|
NM_000683
|
ADRA2C
|
adrenergic, alpha-2C-, receptor
|
|
chr3_+_171619346
|
0.903
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr13_+_95541093
|
0.903
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr4_+_7245218
|
0.900
|
NM_020777
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2
|
|
chr3_-_124228999
|
0.899
|
|
SEMA5B
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B
|
|
chr10_+_115793795
|
0.899
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr6_+_72653126
|
0.895
|
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr4_+_177224142
|
0.895
|
|
WDR17
|
WD repeat domain 17
|
|
chr18_+_53170718
|
0.891
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr10_-_135000464
|
0.890
|
NM_015722
|
CALY
|
calcyon neuron-specific vesicular protein
|
|
chr2_-_42573980
|
0.890
|
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr19_-_7845325
|
0.887
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr4_-_66218777
|
0.881
|
|
EPHA5
|
EPH receptor A5
|
|
chr11_-_18769703
|
0.876
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr4_-_41849333
|
0.875
|
|
BEND4
|
BEN domain containing 4
|
|
chr4_-_5942818
|
0.873
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr13_+_27264779
|
0.871
|
NM_145657
|
GSX1
|
GS homeobox 1
|
|
chr14_-_37795001
|
0.871
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr4_+_147779492
|
0.869
|
NM_004575
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr7_+_45580704
|
0.865
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr17_+_56831951
|
0.862
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr12_-_61614930
|
0.861
|
NM_020700
|
PPM1H
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H
|
|
chr11_-_44288188
|
0.860
|
NM_021926
|
ALX4
|
ALX homeobox 4
|
|
chr2_+_231610438
|
0.852
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr8_+_21937533
|
0.852
|
|
NPM2
|
nucleophosmin/nucleoplasmin 2
|
|
chr2_-_278181
|
0.850
|
NM_001002919
|
FAM150B
|
family with sequence similarity 150, member B
|
|
chr17_-_64108529
|
0.850
|
NM_017565
|
FAM20A
|
family with sequence similarity 20, member A
|
|
chr4_+_93444572
|
0.841
|
NM_001510
|
GRID2
|
glutamate receptor, ionotropic, delta 2
|
|
chr5_+_80292213
|
0.834
|
|
RASGRF2
|
Ras protein-specific guanine nucleotide-releasing factor 2
|
|
chr3_+_182912769
|
0.827
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr10_-_135088065
|
0.827
|
NM_001012508
|
SPRN
|
shadow of prion protein homolog (zebrafish)
|
|
chr19_-_54267932
|
0.822
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr14_+_28305922
|
0.821
|
NM_005249
|
FOXG1
|
forkhead box G1
|
|
chr5_-_134397740
|
0.821
|
NM_002653
|
PITX1
|
paired-like homeodomain 1
|
|
chr20_-_42872295
|
0.820
|
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr15_-_76700563
|
0.819
|
NM_000743
NM_001166694
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3
|
|
chr9_+_967386
|
0.818
|
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chrX_-_74062005
|
0.818
|
NM_001008537
|
KIAA2022
|
KIAA2022
|
|
chr10_+_105334704
|
0.817
|
|
NEURL
|
neuralized homolog (Drosophila)
|
|
chr16_+_1143738
|
0.815
|
|
CACNA1H
|
calcium channel, voltage-dependent, T type, alpha 1H subunit
|