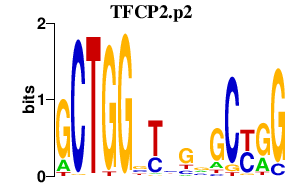
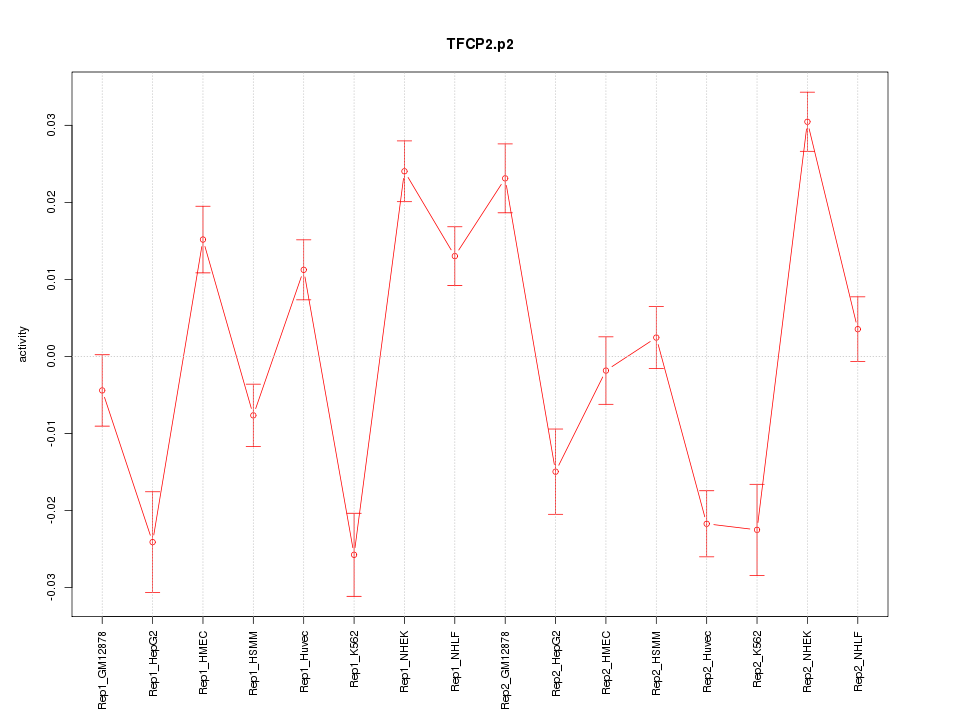
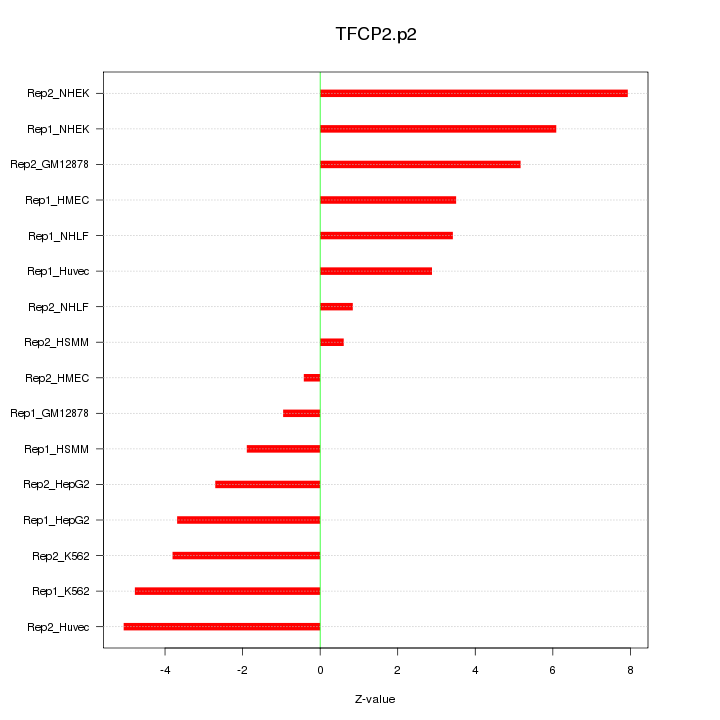
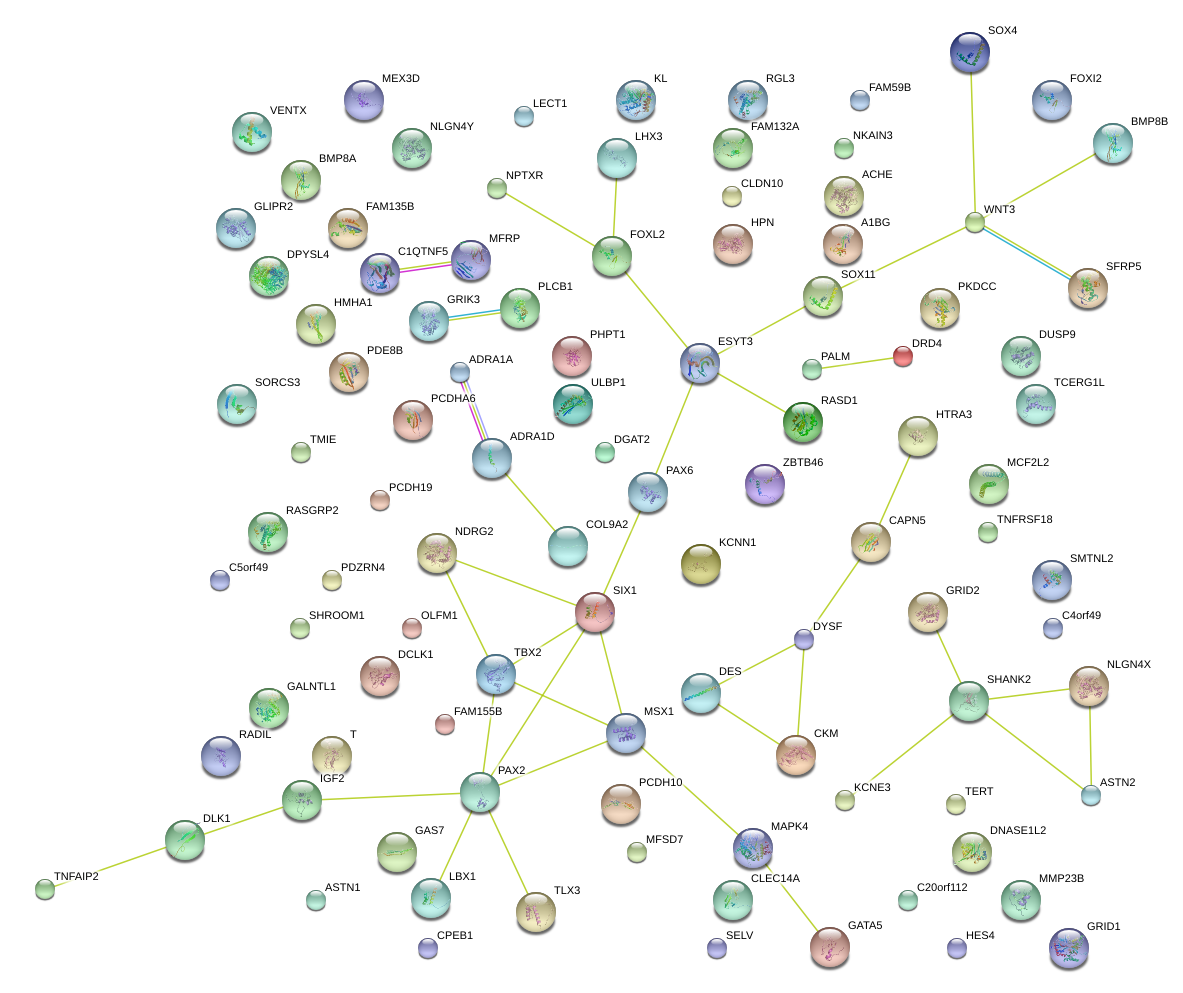
|
chr9_-_138234707
|
6.318
|
NM_014564
|
LHX3
|
LIM homeobox 3
|
|
chr5_+_76542461
|
4.395
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr1_-_40027097
|
4.208
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr1_-_40027108
|
4.160
|
NM_001720
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr8_-_26778832
|
4.127
|
NM_000680
NM_033302
NM_033303
NM_033304
|
ADRA1A
|
adrenergic, alpha-1A-, receptor
|
|
chr13_+_32488477
|
3.970
|
NM_004795
|
KL
|
klotho
|
|
chr10_+_106391032
|
3.883
|
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr5_+_170668892
|
3.852
|
NM_021025
|
TLX3
|
T-cell leukemia homeobox 3
|
|
chr19_-_11390948
|
3.742
|
NM_001035223
NM_001161616
|
RGL3
|
ral guanine nucleotide dissociation stimulator-like 3
|
|
chr7_-_100331417
|
3.712
|
NM_000665
NM_015831
|
ACHE
|
acetylcholinesterase
|
|
chr4_-_672915
|
3.606
|
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr3_-_62835601
|
3.547
|
|
|
|
|
chr3_+_129690735
|
3.393
|
|
|
|
|
chr19_+_17923110
|
3.372
|
NM_002248
|
KCNN1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1
|
|
chr4_-_672967
|
3.354
|
NM_032219
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr10_+_106390848
|
3.301
|
NM_014978
|
SORCS3
|
sortilin-related VPS10 domain containing receptor 3
|
|
chr1_-_40555413
|
3.264
|
|
COL9A2
|
collagen, type IX, alpha 2
|
|
chr20_+_8060762
|
3.101
|
NM_015192
NM_182734
|
PLCB1
|
phospholipase C, beta 1 (phosphoinositide-specific)
|
|
chr10_-_88116181
|
3.053
|
NM_017551
|
GRID1
|
glutamate receptor, ionotropic, delta 1
|
|
chr9_-_119217147
|
3.025
|
|
ASTN2
|
astrotactin 2
|
|
chr17_-_17340219
|
2.878
|
NM_016084
|
RASD1
|
RAS, dexamethasone-induced 1
|
|
chr13_-_52211925
|
2.858
|
NM_001011705
NM_007015
|
LECT1
|
leukocyte cell derived chemotaxin 1
|
|
chr4_+_4912272
|
2.802
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr1_+_1557419
|
2.787
|
NM_006983
|
MMP23B
|
matrix metallopeptidase 23B
|
|
chr1_-_1131795
|
2.771
|
NM_004195
NM_148901
NM_148902
|
TNFRSF18
|
tumor necrosis factor receptor superfamily, member 18
|
|
chr17_+_56831951
|
2.750
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr11_+_627268
|
2.745
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr1_-_925332
|
2.698
|
NM_001142467
NM_021170
|
HES4
|
hairy and enhancer of split 4 (Drosophila)
|
|
chr2_+_42128521
|
2.681
|
NM_138370
|
PKDCC
|
protein kinase domain containing, cytoplasmic homolog (mouse)
|
|
chr10_+_134901397
|
2.671
|
NM_014468
|
VENTX
|
VENT homeobox homolog (Xenopus laevis)
|
|
chr11_-_73856241
|
2.667
|
NM_005472
|
KCNE3
|
potassium voltage-gated channel, Isk-related family, member 3
|
|
chr16_+_2226468
|
2.660
|
NM_001374
|
DNASE1L2
|
deoxyribonuclease I-like 2
|
|
chr22_-_37569962
|
2.622
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr12_+_39868434
|
2.600
|
NM_001164595
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chr5_+_140286485
|
2.586
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr9_-_119217128
|
2.564
|
NM_014010
|
ASTN2
|
astrotactin 2
|
|
chr20_-_30636535
|
2.501
|
|
LOC284804
|
hypothetical protein LOC284804
|
|
chr1_-_37272295
|
2.466
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr19_+_44697592
|
2.439
|
NM_182704
|
SELV
|
selenoprotein V
|
|
chr8_-_139578246
|
2.423
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr11_-_70641267
|
2.412
|
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr2_+_26249463
|
2.398
|
NM_001168241
|
FAM59B
|
family with sequence similarity 59, member B
|
|
chr17_+_56832255
|
2.374
|
|
TBX2
|
T-box 2
|
|
chr14_+_68796621
|
2.369
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr15_-_81113676
|
2.366
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr5_-_1348103
|
2.356
|
NM_001193376
NM_198253
|
TERT
|
telomerase reverse transcriptase
|
|
chr14_-_60185659
|
2.352
|
NM_005982
|
SIX1
|
SIX homeobox 1
|
|
chr14_-_37795001
|
2.346
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr19_-_63550712
|
2.335
|
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr5_-_132189762
|
2.332
|
NM_133456
|
SHROOM1
|
shroom family member 1
|
|
chr11_-_31796061
|
2.327
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr1_-_175400573
|
2.326
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr9_+_137107222
|
2.253
|
|
OLFM1
|
olfactomedin 1
|
|
chr2_+_71534260
|
2.243
|
NM_001130976
NM_001130977
NM_001130978
NM_001130979
NM_001130980
NM_001130981
NM_003494
|
DYSF
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive)
|
|
chr11_-_2116048
|
2.232
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr19_+_1022208
|
2.229
|
|
HMHA1
|
histocompatibility (minor) HA-1
|
|
chr19_-_1518875
|
2.228
|
NM_001174118
NM_203304
|
MEX3D
|
mex-3 homolog D (C. elegans)
|
|
chr7_-_4889772
|
2.209
|
NM_018059
|
RADIL
|
Ras association and DIL domains
|
|
chr17_-_10042579
|
2.198
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chrX_+_152565849
|
2.195
|
|
DUSP9
|
dual specificity phosphatase 9
|
|
chr1_-_175400665
|
2.195
|
|
ASTN1
|
astrotactin 1
|
|
chr19_+_659766
|
2.186
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr10_+_133850325
|
2.181
|
NM_006426
|
DPYSL4
|
dihydropyrimidinase-like 4
|
|
chrX_-_99551926
|
2.156
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr22_-_37569679
|
2.149
|
|
|
|
|
chr10_-_132999798
|
2.139
|
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr6_-_166502119
|
2.101
|
NM_003181
|
T
|
T, brachyury homolog (mouse)
|
|
chr9_+_138862212
|
2.098
|
|
PHPT1
|
phosphohistidine phosphatase 1
|
|
chr3_+_46717826
|
2.060
|
NM_147196
|
TMIE
|
transmembrane inner ear
|
|
chr20_-_60484420
|
2.050
|
NM_080473
|
GATA5
|
GATA binding protein 5
|
|
chrX_+_68641802
|
2.049
|
NM_015686
|
FAM155B
|
family with sequence similarity 155, member B
|
|
chr13_-_35603466
|
2.039
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr13_-_35603432
|
2.030
|
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr6_+_150326802
|
2.027
|
NM_025218
|
ULBP1
|
UL16 binding protein 1
|
|
chr10_+_102495327
|
2.019
|
NM_000278
NM_003987
NM_003988
NM_003989
NM_003990
|
PAX2
|
paired box 2
|
|
chr3_+_139636111
|
2.014
|
|
ESYT3
|
extended synaptotagmin-like protein 3
|
|
chr1_-_1171964
|
2.011
|
NM_001014980
|
FAM132A
|
family with sequence similarity 132, member A
|
|
chr9_+_36126667
|
2.010
|
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr13_+_95002942
|
2.001
|
NM_006984
|
CLDN10
|
claudin 10
|
|
chr17_+_56832492
|
1.998
|
|
TBX2
|
T-box 2
|
|
chr9_+_36126710
|
1.992
|
NM_022343
|
GLIPR2
|
GLI pathogenesis-related 2
|
|
chr10_-_99521701
|
1.992
|
NM_003015
|
SFRP5
|
secreted frizzled-related protein 5
|
|
chr11_-_2116360
|
1.990
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_+_214668
|
1.990
|
|
|
|
|
chr11_-_118716736
|
1.984
|
|
C1QTNF5
MFRP
|
C1q and tumor necrosis factor related protein 5
membrane frizzled-related protein
|
|
chr11_-_64268841
|
1.977
|
NM_001098670
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr10_-_102978678
|
1.975
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr4_-_140420789
|
1.973
|
NM_032623
|
C4orf49
|
chromosome 4 open reading frame 49
|
|
chr4_+_93444572
|
1.971
|
NM_001510
|
GRID2
|
glutamate receptor, ionotropic, delta 2
|
|
chr8_+_63324053
|
1.970
|
NM_173688
|
NKAIN3
|
Na+/K+ transporting ATPase interacting 3
|
|
chr3_+_139636104
|
1.967
|
NM_031913
|
ESYT3
|
extended synaptotagmin-like protein 3
|
|
chr3_+_129682434
|
1.955
|
|
|
|
|
chr3_-_184628548
|
1.915
|
NM_015078
|
MCF2L2
|
MCF.2 cell line derived transforming sequence-like 2
|
|
chr1_-_40026718
|
1.905
|
|
BMP8B
|
bone morphogenetic protein 8b
|
|
chr17_-_60208083
|
1.901
|
|
LOC146880
|
hypothetical LOC146880
|
|
chr14_-_20562962
|
1.887
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr9_+_137107088
|
1.869
|
|
OLFM1
|
olfactomedin 1
|
|
chr2_+_5750229
|
1.864
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr5_-_7904255
|
1.856
|
NM_001089584
|
C5orf49
|
chromosome 5 open reading frame 49
|
|
chr10_-_132999973
|
1.843
|
NM_174937
|
TCERG1L
|
transcription elongation regulator 1-like
|
|
chr19_+_40223249
|
1.841
|
NM_002151
NM_182983
|
HPN
|
hepsin
|
|
chr4_+_8322358
|
1.839
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr17_+_4434582
|
1.810
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr19_-_50518072
|
1.807
|
NM_001824
|
CKM
|
creatine kinase, muscle
|
|
chr18_+_46340400
|
1.802
|
NM_002747
|
MAPK4
|
mitogen-activated protein kinase 4
|
|
chr20_-_61933012
|
1.800
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chrX_+_128942435
|
1.779
|
|
|
|
|
chr11_-_118716244
|
1.771
|
|
MFRP
|
membrane frizzled-related protein
|
|
chr14_+_102659464
|
1.762
|
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr2_+_219991342
|
1.760
|
NM_001927
|
DES
|
desmin
|
|
chr3_-_140148671
|
1.757
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr9_+_137107187
|
1.755
|
|
OLFM1
|
olfactomedin 1
|
|
chr10_+_129425527
|
1.745
|
NM_207426
|
FOXI2
|
forkhead box I2
|
|
chr11_+_75157414
|
1.741
|
NM_032564
|
DGAT2
|
diacylglycerol O-acyltransferase 2
|
|
chr13_-_35603513
|
1.728
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr17_-_42250961
|
1.725
|
NM_030753
|
WNT3
|
wingless-type MMTV integration site family, member 3
|
|
chr4_-_673107
|
1.724
|
|
MFSD7
|
major facilitator superfamily domain containing 7
|
|
chr14_+_100263066
|
1.719
|
|
DLK1
|
delta-like 1 homolog (Drosophila)
|
|
chrX_-_6155887
|
1.710
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr19_-_38408449
|
1.709
|
NM_019849
|
SLC7A10
|
solute carrier family 7, (neutral amino acid transporter, y+ system) member 10
|
|
chr8_-_53640520
|
1.702
|
NM_207413
|
FAM150A
|
family with sequence similarity 150, member A
|
|
chr15_-_56145140
|
1.691
|
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr19_-_38485161
|
1.688
|
|
|
|
|
chr17_+_42283946
|
1.686
|
NM_003396
|
WNT9B
|
wingless-type MMTV integration site family, member 9B
|
|
chr14_+_30413393
|
1.681
|
NM_001135058
NM_004086
|
COCH
|
coagulation factor C homolog, cochlin (Limulus polyphemus)
|
|
chr19_+_38376980
|
1.678
|
|
LRP3
|
low density lipoprotein receptor-related protein 3
|
|
chr16_+_1323627
|
1.678
|
|
BAIAP3
|
BAI1-associated protein 3
|
|
chr1_+_200124412
|
1.667
|
NM_198149
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr5_-_115938305
|
1.661
|
NM_020796
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A
|
|
chr17_+_45404677
|
1.657
|
|
DLX4
|
distal-less homeobox 4
|
|
chr9_+_91409746
|
1.642
|
NM_006705
|
GADD45G
|
growth arrest and DNA-damage-inducible, gamma
|
|
chr9_-_138757175
|
1.637
|
NM_001001712
|
LCN10
|
lipocalin 10
|
|
chr16_+_82559600
|
1.631
|
NM_019065
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2
|
|
chr22_+_22445035
|
1.615
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr13_+_112681626
|
1.612
|
NM_024979
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr9_-_103540038
|
1.607
|
|
GRIN3A
|
glutamate receptor, ionotropic, N-methyl-D-aspartate 3A
|
|
chrX_-_106846684
|
1.600
|
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr9_-_139455611
|
1.593
|
NM_001033113
NM_198585
|
ENTPD8
|
ectonucleoside triphosphate diphosphohydrolase 8
|
|
chr10_-_104158910
|
1.586
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr17_+_42686153
|
1.574
|
NM_000212
|
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61)
|
|
chr17_+_42686248
|
1.574
|
|
|
|
|
chr8_+_16929116
|
1.572
|
NM_181723
|
EFHA2
|
EF-hand domain family, member A2
|
|
chr11_+_110675175
|
1.569
|
NM_001136105
|
C11orf93
|
chromosome 11 open reading frame 93
|
|
chr11_+_61861458
|
1.565
|
|
ASRGL1
|
asparaginase like 1
|
|
chr11_+_61861349
|
1.562
|
NM_001083926
NM_025080
|
ASRGL1
|
asparaginase like 1
|
|
chr11_-_2116501
|
1.557
|
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr12_-_56418295
|
1.555
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr1_-_6162669
|
1.552
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr14_-_51605487
|
1.552
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr2_-_236741352
|
1.548
|
NM_001485
|
GBX2
|
gastrulation brain homeobox 2
|
|
chr11_-_64266916
|
1.545
|
|
|
|
|
chr15_-_91433165
|
1.542
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr14_-_99842519
|
1.540
|
NM_001039355
|
SLC25A29
|
solute carrier family 25, member 29
|
|
chr7_-_100630912
|
1.529
|
NM_178176
|
MOGAT3
|
monoacylglycerol O-acyltransferase 3
|
|
chr9_+_137106909
|
1.529
|
NM_014279
NM_006334
|
OLFM1
|
olfactomedin 1
|
|
chr9_+_8848130
|
1.523
|
|
|
|
|
chr11_+_2116486
|
1.520
|
|
|
|
|
chr9_+_125813849
|
1.519
|
|
LHX2
|
LIM homeobox 2
|
|
chr7_+_128257698
|
1.514
|
NM_001127487
NM_001458
|
FLNC
|
filamin C, gamma
|
|
chr9_-_138762725
|
1.510
|
NM_198946
|
LCN6
|
lipocalin 6
|
|
chr1_+_95355415
|
1.507
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr4_-_1156458
|
1.501
|
|
SPON2
|
spondin 2, extracellular matrix protein
|
|
chr16_+_3036682
|
1.497
|
NM_022468
|
MMP25
|
matrix metallopeptidase 25
|
|
chr17_-_3814334
|
1.496
|
NM_005173
NM_174953
NM_174954
NM_174955
NM_174956
NM_174957
NM_174958
|
ATP2A3
|
ATPase, Ca++ transporting, ubiquitous
|
|
chr3_+_148610516
|
1.493
|
|
ZIC1
|
Zic family member 1 (odd-paired homolog, Drosophila)
|
|
chr9_+_137107007
|
1.492
|
|
OLFM1
|
olfactomedin 1
|
|
chr11_-_2116775
|
1.491
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr3_+_127743868
|
1.491
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr1_+_200124620
|
1.487
|
|
SHISA4
|
shisa homolog 4 (Xenopus laevis)
|
|
chr11_-_407324
|
1.478
|
NM_001135053
NM_021805
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain
|
|
chr1_-_152741148
|
1.471
|
NM_001010846
|
SHE
|
Src homology 2 domain containing E
|
|
chr9_+_966963
|
1.471
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr11_-_31782169
|
1.470
|
|
PAX6
|
paired box 6
|
|
chr20_-_26137755
|
1.467
|
|
LOC284801
|
hypothetical protein LOC284801
|
|
chr13_-_49597776
|
1.466
|
|
DLEU2
|
deleted in lymphocytic leukemia 2 (non-protein coding)
|
|
chr22_-_37180958
|
1.459
|
NM_152868
|
KCNJ4
|
potassium inwardly-rectifying channel, subfamily J, member 4
|
|
chr11_-_16992524
|
1.458
|
NM_175058
|
PLEKHA7
|
pleckstrin homology domain containing, family A member 7
|
|
chr11_+_65311142
|
1.457
|
|
OVOL1
|
ovo-like 1(Drosophila)
|
|
chr7_-_44331544
|
1.449
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr1_-_1465602
|
1.446
|
NM_001114748
|
C1orf70
|
chromosome 1 open reading frame 70
|
|
chr1_+_41022066
|
1.446
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr3_-_47595190
|
1.443
|
NM_006574
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C)
|
|
chr11_-_118799452
|
1.441
|
NM_006288
|
THY1
|
Thy-1 cell surface antigen
|
|
chr3_-_22389126
|
1.436
|
|
ZNF385D
|
zinc finger protein 385D
|
|
chrX_-_132377087
|
1.434
|
|
GPC4
|
glypican 4
|
|
chr14_+_105024301
|
1.431
|
NM_001311
|
CRIP1
|
cysteine-rich protein 1 (intestinal)
|
|
chr2_+_220087135
|
1.426
|
NM_018674
NM_182847
|
ACCN4
|
amiloride-sensitive cation channel 4, pituitary
|
|
chr14_-_51605461
|
1.425
|
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr6_+_99389300
|
1.421
|
NM_005604
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr10_+_134823925
|
1.416
|
NM_152643
|
KNDC1
|
kinase non-catalytic C-lobe domain (KIND) containing 1
|
|
chr4_-_110443247
|
1.411
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr1_+_39729885
|
1.405
|
NM_181809
|
BMP8A
|
bone morphogenetic protein 8a
|
|
chr12_+_5023345
|
1.405
|
NM_002234
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5
|
|
chr14_+_28306633
|
1.404
|
|
FOXG1
|
forkhead box G1
|
|
chr17_+_42686217
|
1.403
|
|
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61)
|
|
chr22_-_30929463
|
1.403
|
NM_001098527
|
RFPL2
|
ret finger protein-like 2
|
|
chr8_+_31616809
|
1.399
|
NM_013962
|
NRG1
|
neuregulin 1
|
|
chr1_+_152741318
|
1.393
|
NM_001098475
NM_182499
|
TDRD10
|
tudor domain containing 10
|
|
chr1_-_25129305
|
1.386
|
|
RUNX3
|
runt-related transcription factor 3
|
|
chr19_+_3129735
|
1.374
|
NM_003775
|
S1PR4
|
sphingosine-1-phosphate receptor 4
|