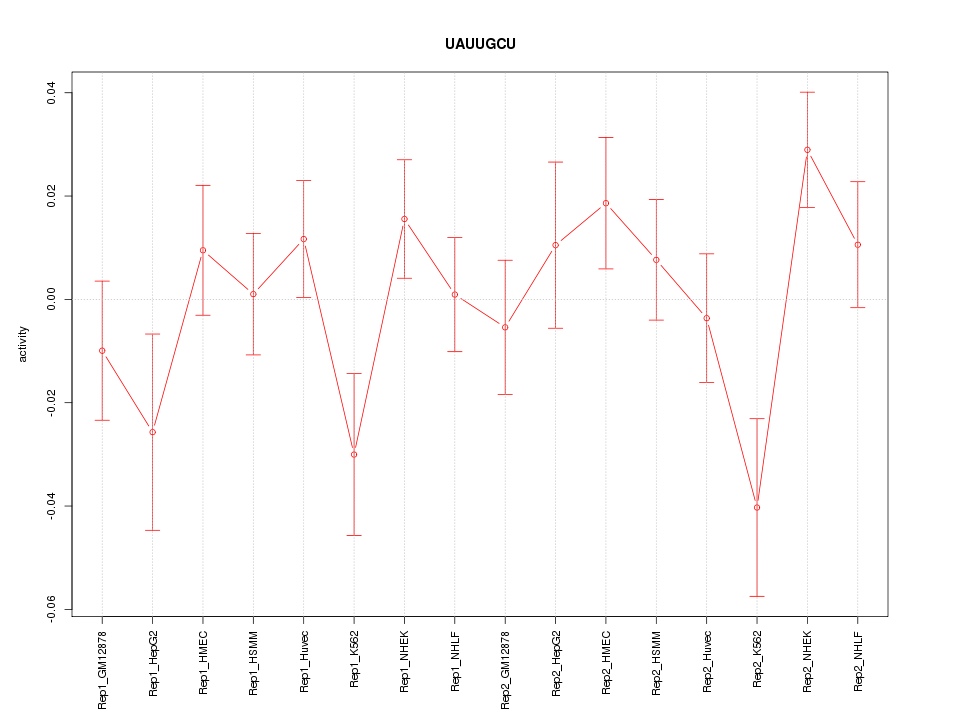
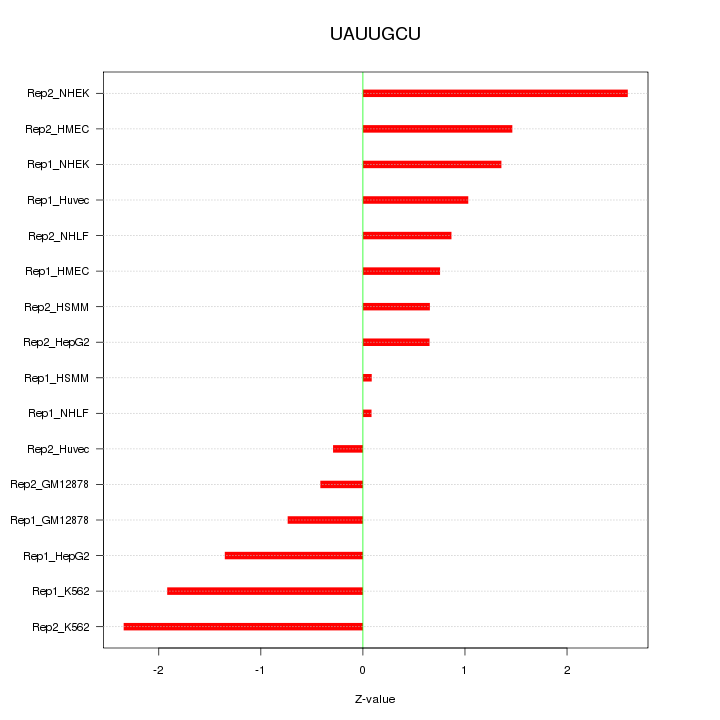
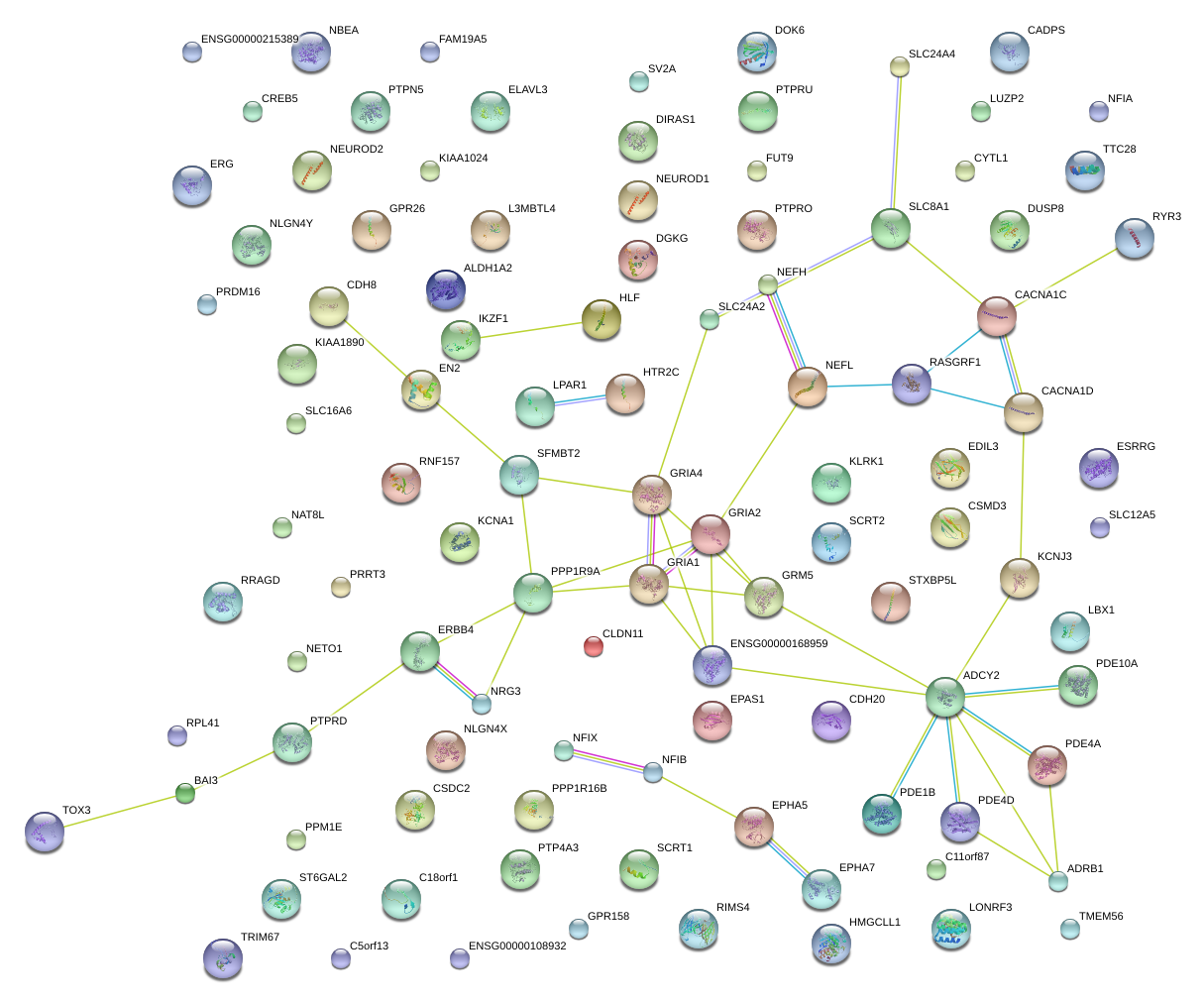
|
chr15_-_56145140
|
2.488
|
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr6_-_94185780
|
2.369
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr10_+_115793795
|
2.188
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr22_+_28206180
|
2.131
|
NM_021076
|
NEFH
|
neurofilament, heavy polypeptide
|
|
chr10_-_7493418
|
2.086
|
NM_001029880
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr12_+_4889309
|
2.016
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr4_-_66218247
|
1.917
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr18_-_68685789
|
1.913
|
NM_138966
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr1_-_214963279
|
1.911
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_-_7491204
|
1.825
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr15_+_31390454
|
1.819
|
NM_001036
|
RYR3
|
ryanodine receptor 3
|
|
chr7_+_94377145
|
1.811
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr7_+_154943466
|
1.802
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr11_+_24475091
|
1.796
|
NM_001009909
|
LUZP2
|
leucine zipper protein 2
|
|
chr19_+_10392128
|
1.755
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_10402334
|
1.744
|
NM_001111308
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr2_-_213111525
|
1.741
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr1_-_215377718
|
1.730
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr15_-_77170135
|
1.704
|
NM_001145648
NM_002891
|
RASGRF1
|
Ras protein-specific guanine nucleotide-releasing factor 1
|
|
chr19_-_11452439
|
1.657
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr10_+_83625049
|
1.650
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr8_-_24869945
|
1.615
|
NM_006158
|
NEFL
|
neurofilament, light polypeptide
|
|
chr18_-_6404900
|
1.596
|
NM_173464
|
L3MBTL4
|
l(3)mbt-like 4 (Drosophila)
|
|
chr10_+_25504295
|
1.595
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr17_-_71747867
|
1.577
|
NM_052916
|
RNF157
|
ring finger protein 157
|
|
chr5_-_59225377
|
1.564
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr18_+_65219114
|
1.538
|
NM_152721
|
DOK6
|
docking protein 6
|
|
chr20_+_36867708
|
1.531
|
NM_001172735
NM_015568
|
PPP1R16B
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chrX_-_6155887
|
1.528
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr19_-_2672337
|
1.512
|
NM_145173
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chrX_-_6156655
|
1.490
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr3_+_122109716
|
1.480
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr20_-_42872295
|
1.453
|
NM_182970
|
RIMS4
|
regulating synaptic membrane exocytosis 4
|
|
chr16_-_60627536
|
1.445
|
NM_001796
|
CDH8
|
cadherin 8, type 2
|
|
chr8_-_114518194
|
1.445
|
NM_052900
NM_198123
|
CSMD3
|
CUB and Sushi multiple domains 3
|
|
chr11_-_18769703
|
1.427
|
NM_001039970
NM_006906
NM_032781
|
PTPN5
|
protein tyrosine phosphatase, non-receptor type 5 (striatum-enriched)
|
|
chr3_-_62836052
|
1.386
|
NM_003716
NM_183393
NM_183394
|
CADPS
|
Ca++-dependent secretion activator
|
|
chrX_+_113724806
|
1.385
|
NM_000868
|
HTR2C
|
5-hydroxytryptamine (serotonin) receptor 2C
|
|
chr6_-_165995510
|
1.382
|
NM_001130690
NM_006661
|
PDE10A
|
phosphodiesterase 10A
|
|
chr1_+_2975590
|
1.364
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr12_+_2032649
|
1.352
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr11_-_1549719
|
1.347
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr16_-_51138306
|
1.295
|
NM_001080430
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr9_-_19778590
|
1.286
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr10_+_125415860
|
1.286
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chr12_+_53241365
|
1.284
|
NM_001165975
|
PDE1B
|
phosphodiesterase 1B, calmodulin-dependent
|
|
chr21_-_38955487
|
1.277
|
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_+_108798047
|
1.273
|
NM_207645
|
C11orf87
|
chromosome 11 open reading frame 87
|
|
chr22_-_27405708
|
1.267
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr9_-_10602722
|
1.259
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr11_-_88436463
|
1.258
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr10_-_102978678
|
1.243
|
NM_006562
|
LBX1
|
ladybird homeobox 1
|
|
chr7_+_50314801
|
1.238
|
NM_006060
|
IKZF1
|
IKAROS family zinc finger 1 (Ikaros)
|
|
chr22_+_47263919
|
1.235
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr17_-_63798851
|
1.232
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr7_+_94374862
|
1.232
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr17_+_50697319
|
1.228
|
NM_002126
|
HLF
|
hepatic leukemia factor
|
|
chr12_+_15366712
|
1.222
|
NM_002848
NM_030667
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr5_+_7449030
|
1.217
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr6_-_55551812
|
1.212
|
NM_001042406
NM_019036
|
HMGCLL1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1
|
|
chr5_-_83716346
|
1.205
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chr3_+_53504070
|
1.203
|
NM_000720
NM_001128839
NM_001128840
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit
|
|
chrX_+_117992623
|
1.187
|
NM_001031855
NM_024778
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr3_+_171619346
|
1.175
|
NM_005602
|
CLDN11
|
claudin 11
|
|
chr1_-_148155986
|
1.172
|
NM_014849
|
SV2A
|
synaptic vesicle glycoprotein 2A
|
|
chr17_+_54188230
|
1.168
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr18_+_13208777
|
1.166
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr22_+_40286957
|
1.165
|
NM_014460
|
CSDC2
|
cold shock domain containing C2, RNA binding
|
|
chr6_-_90178685
|
1.160
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr1_+_95355415
|
1.156
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr2_-_182253457
|
1.090
|
NM_002500
|
NEUROD1
|
neurogenic differentiation 1
|
|
chr11_+_104986009
|
1.088
|
NM_000829
NM_001077243
NM_001077244
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr5_-_111340421
|
1.083
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr16_-_51139214
|
1.082
|
NM_001146188
|
TOX3
|
TOX high mobility group box family member 3
|
|
chr15_+_77511912
|
1.079
|
NM_015206
|
KIAA1024
|
KIAA1024
|
|
chr1_+_229365296
|
1.076
|
NM_001004342
|
TRIM67
|
tripartite motif containing 67
|
|
chr2_+_155263338
|
1.060
|
NM_002239
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3
|
|
chr18_+_57308754
|
1.055
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr20_+_44091219
|
1.053
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr13_+_34414390
|
1.053
|
NM_015678
|
NBEA
|
neurobeachin
|
|
chr19_+_10424581
|
1.047
|
NM_006202
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr3_-_187562470
|
1.046
|
NM_001080744
NM_001080745
NM_001346
|
DGKG
|
diacylglycerol kinase, gamma 90kDa
|
|
chr10_+_83627422
|
1.042
|
NM_001165973
|
NRG3
|
neuregulin 3
|
|
chr17_-_35017691
|
1.035
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr1_-_215329569
|
1.031
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr8_-_4839670
|
1.013
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr6_+_96570565
|
1.009
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr8_+_142501188
|
1.007
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr4_+_2031036
|
0.997
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr20_-_604822
|
0.992
|
NM_033129
|
SCRT2
|
scratch homolog 2, zinc finger protein (Drosophila)
|
|
chr2_-_106869041
|
0.982
|
NM_001142351
|
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2
|
|
chr7_+_28441758
|
0.971
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_-_8723893
|
0.968
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr2_-_40593074
|
0.967
|
NM_001112802
|
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1
|
|
chr6_+_69401660
|
0.964
|
NM_001704
|
BAI3
|
brain-specific angiogenesis inhibitor 3
|
|
chr9_-_14388981
|
0.961
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chrX_-_106248706
|
0.960
|
NM_001171080
NM_018301
|
RBM41
|
RNA binding motif protein 41
|
|
chr3_+_11009419
|
0.957
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr17_-_37590910
|
0.945
|
NM_001524
|
HCRT
|
hypocretin (orexin) neuropeptide precursor
|
|
chr2_-_217268492
|
0.930
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr19_+_540849
|
0.929
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr22_+_48951286
|
0.919
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr2_+_5750229
|
0.908
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr1_-_46370837
|
0.900
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr21_-_26867138
|
0.887
|
NM_052954
|
CYYR1
|
cysteine/tyrosine-rich 1
|
|
chr4_+_41309668
|
0.883
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr22_+_47350781
|
0.877
|
NM_015381
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr16_+_65436341
|
0.876
|
NM_001014435
|
CA7
|
carbonic anhydrase VII
|
|
chr12_+_2774362
|
0.875
|
NM_002014
|
FKBP4
|
FK506 binding protein 4, 59kDa
|
|
chr3_+_185430910
|
0.866
|
NM_138345
|
VWA5B2
|
von Willebrand factor A domain containing 5B2
|
|
chr11_+_91724909
|
0.861
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr20_-_3097070
|
0.861
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chr16_-_10184111
|
0.859
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr1_+_235272324
|
0.845
|
NM_001035
|
RYR2
|
ryanodine receptor 2 (cardiac)
|
|
chr20_+_24397834
|
0.837
|
NM_024893
|
TMEM90B
|
transmembrane protein 90B
|
|
chr19_-_7845325
|
0.834
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr7_-_19123787
|
0.814
|
NM_000474
|
TWIST1
|
twist homolog 1 (Drosophila)
|
|
chr12_+_77781903
|
0.807
|
NM_001135805
|
SYT1
|
synaptotagmin I
|
|
chr6_-_84475827
|
0.786
|
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr11_-_132318080
|
0.784
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr7_-_123460722
|
0.781
|
NM_001136002
|
TMEM229A
|
transmembrane protein 229A
|
|
chr2_+_176689737
|
0.765
|
NM_002148
|
HOXD10
|
homeobox D10
|
|
chr15_+_83160914
|
0.763
|
NM_020778
|
ALPK3
|
alpha-kinase 3
|
|
chr19_+_45664889
|
0.754
|
NM_020971
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4
|
|
chr14_+_20319049
|
0.753
|
NM_005615
|
RNASE6
|
ribonuclease, RNase A family, k6
|
|
chr9_-_92444886
|
0.747
|
NM_017594
|
DIRAS2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr16_+_28211123
|
0.743
|
NM_001024401
|
SBK1
|
SH3-binding domain kinase 1
|
|
chr14_+_92459197
|
0.741
|
NM_001275
|
CHGA
|
chromogranin A (parathyroid secretory protein 1)
|
|
chr3_-_127558747
|
0.737
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr8_-_145530639
|
0.735
|
NM_031309
|
SCRT1
|
scratch homolog 1, zinc finger protein (Drosophila)
|
|
chr11_-_106394039
|
0.728
|
NM_000855
|
GUCY1A2
|
guanylate cyclase 1, soluble, alpha 2
|
|
chr9_+_966963
|
0.725
|
NM_021240
|
DMRT3
|
doublesex and mab-3 related transcription factor 3
|
|
chr13_-_20374895
|
0.722
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr5_-_58688428
|
0.707
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr22_+_38296703
|
0.704
|
NM_001003406
NM_021096
|
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit
|
|
chr17_+_45993338
|
0.698
|
NM_018896
NM_198376
NM_198377
NM_198378
NM_198379
NM_198380
NM_198382
NM_198383
NM_198384
NM_198385
NM_198386
NM_198387
NM_198388
NM_198396
NM_198397
|
CACNA1G
|
calcium channel, voltage-dependent, T type, alpha 1G subunit
|
|
chr22_-_31784328
|
0.697
|
NM_001135774
|
SYN3
|
synapsin III
|
|
chr10_-_71811353
|
0.694
|
NM_207119
|
LRRC20
|
leucine rich repeat containing 20
|
|
chr11_-_63440891
|
0.694
|
NM_173587
|
RCOR2
|
REST corepressor 2
|
|
chr3_+_179736917
|
0.693
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr3_-_13089547
|
0.690
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr12_-_9804682
|
0.684
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr3_+_171621721
|
0.682
|
NM_001185056
|
CLDN11
|
claudin 11
|
|
chr4_+_41057507
|
0.682
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr7_+_28418668
|
0.680
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr6_+_126112417
|
0.677
|
NM_012259
|
HEY2
|
hairy/enhancer-of-split related with YRPW motif 2
|
|
chr1_+_234916376
|
0.677
|
NM_001103
|
ACTN2
|
actinin, alpha 2
|
|
chr11_-_66092534
|
0.661
|
NM_003793
|
CTSF
|
cathepsin F
|
|
chr14_-_26136799
|
0.661
|
NM_002515
NM_006489
NM_006491
|
NOVA1
|
neuro-oncological ventral antigen 1
|
|
chr10_-_102269580
|
0.655
|
NM_015490
|
SEC31B
|
SEC31 homolog B (S. cerevisiae)
|
|
chr19_+_10404110
|
0.649
|
NM_001111309
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr19_+_1236766
|
0.646
|
NM_001405
|
EFNA2
|
ephrin-A2
|
|
chr5_+_119827903
|
0.642
|
NM_016644
|
PRR16
|
proline rich 16
|
|
chr3_+_12020803
|
0.642
|
NM_003178
NM_133625
|
SYN2
|
synapsin II
|
|
chr11_-_70185559
|
0.641
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr8_-_29262240
|
0.640
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr17_-_34635446
|
0.626
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr20_-_55275078
|
0.625
|
NM_001719
|
BMP7
|
bone morphogenetic protein 7
|
|
chr10_+_35575958
|
0.618
|
NM_181698
|
CCNY
|
cyclin Y
|
|
chr10_-_64246129
|
0.611
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr4_-_141568210
|
0.606
|
NM_001130675
NM_004362
|
CLGN
|
calmegin
|
|
chr1_-_37272295
|
0.604
|
NM_000831
|
GRIK3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr2_+_107969410
|
0.595
|
NM_021815
|
SLC5A7
|
solute carrier family 5 (choline transporter), member 7
|
|
chr1_+_153317887
|
0.592
|
NM_004952
|
EFNA3
|
ephrin-A3
|
|
chr9_-_90983501
|
0.586
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr12_+_63849606
|
0.582
|
NM_001167614
NM_014319
|
LEMD3
|
LEM domain containing 3
|
|
chr5_+_161209969
|
0.572
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr3_+_51716120
|
0.572
|
NM_000839
NM_001130063
|
GRM2
|
glutamate receptor, metabotropic 2
|
|
chr17_-_1412774
|
0.564
|
NM_006224
|
PITPNA
|
phosphatidylinositol transfer protein, alpha
|
|
chr14_+_78815406
|
0.563
|
NM_001105250
NM_138970
|
NRXN3
|
neurexin 3
|
|
chr16_-_10183616
|
0.559
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr2_+_185171337
|
0.555
|
NM_194250
|
ZNF804A
|
zinc finger protein 804A
|
|
chr19_+_7365964
|
0.545
|
NM_015318
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr5_-_153838013
|
0.545
|
NM_004821
|
HAND1
|
heart and neural crest derivatives expressed 1
|
|
chr2_-_158162271
|
0.543
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr16_-_10183244
|
0.542
|
NM_001134408
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr17_-_1478859
|
0.536
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr1_-_119333701
|
0.533
|
NM_152380
|
TBX15
|
T-box 15
|
|
chr20_+_48032918
|
0.533
|
NM_005985
|
SNAI1
|
snail homolog 1 (Drosophila)
|
|
chr20_+_19141282
|
0.532
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr3_+_151803755
|
0.529
|
NM_016275
|
SELT
|
selenoprotein T
|
|
chr2_-_100400481
|
0.526
|
NM_004854
|
CHST10
|
carbohydrate sulfotransferase 10
|
|
chr22_+_23753940
|
0.523
|
NM_001145206
|
KIAA1671
|
KIAA1671
|
|
chr11_+_74629597
|
0.522
|
NM_001195528
|
LOC441617
|
hypothetical LOC441617
|
|
chr3_+_49566901
|
0.522
|
NM_003458
|
BSN
|
bassoon (presynaptic cytomatrix protein)
|
|
chr14_+_71469538
|
0.521
|
NM_004296
|
RGS6
|
regulator of G-protein signaling 6
|
|
chr4_+_55218663
|
0.519
|
NM_000222
NM_001093772
|
KIT
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
|
|
chr13_-_35603513
|
0.518
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr8_-_139578246
|
0.515
|
NM_015912
|
FAM135B
|
family with sequence similarity 135, member B
|
|
chr16_-_10943716
|
0.511
|
NM_014015
|
DEXI
|
Dexi homolog (mouse)
|
|
chr2_-_50428395
|
0.507
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr7_-_44071640
|
0.506
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr11_-_30564338
|
0.505
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chrX_-_39841599
|
0.501
|
NM_001123385
NM_017745
|
BCOR
|
BCL6 corepressor
|
|
chr14_+_74968589
|
0.498
|
NM_001135049
|
JDP2
|
Jun dimerization protein 2
|
|
chr5_+_15553288
|
0.496
|
NM_012304
|
FBXL7
|
F-box and leucine-rich repeat protein 7
|
|
chr8_+_106400322
|
0.494
|
NM_012082
|
ZFPM2
|
zinc finger protein, multitype 2
|
|
chr7_+_45580606
|
0.487
|
NM_021116
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr4_-_135342352
|
0.486
|
NM_001114734
|
PABPC4L
|
poly(A) binding protein, cytoplasmic 4-like
|
|
chr2_-_200044233
|
0.483
|
NM_015265
|
SATB2
|
SATB homeobox 2
|