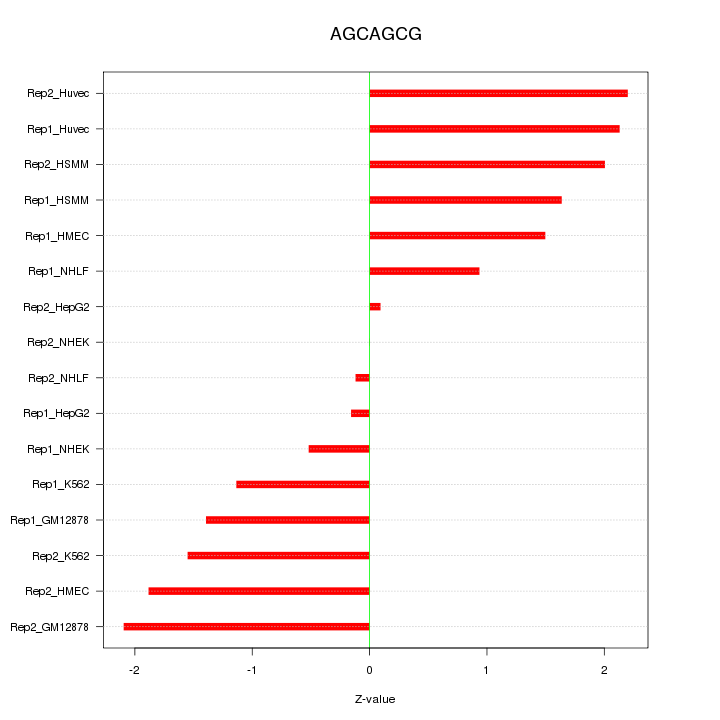
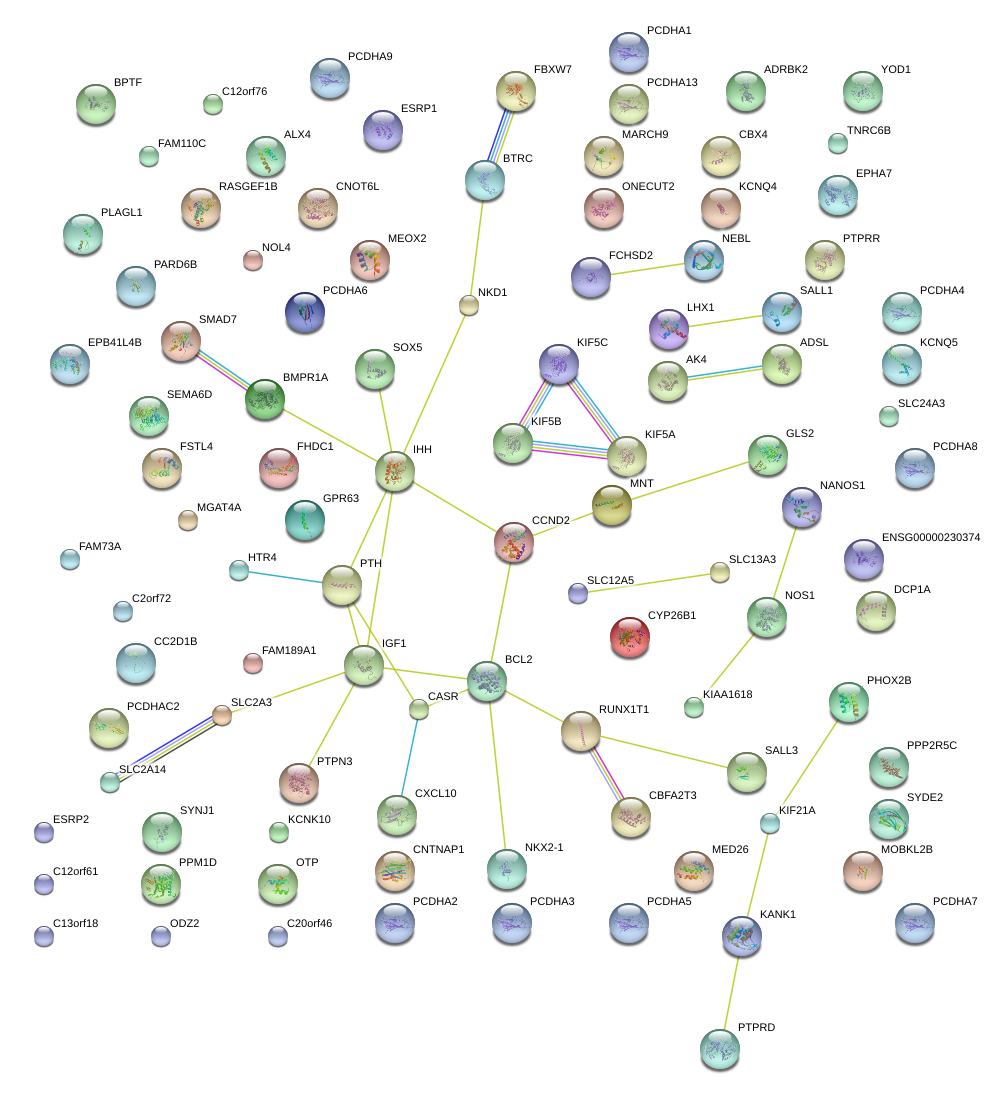
|
chr12_+_4253143
|
4.218
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr9_-_27519434
|
3.158
|
NM_024761
|
MOBKL2B
|
MOB1, Mps One Binder kinase activator-like 2B (yeast)
|
|
chr6_-_144427340
|
2.590
|
NM_001080951
NM_001080955
|
PLAGL1
|
pleiomorphic adenoma gene-like 1
|
|
chr20_+_48781457
|
2.536
|
NM_032521
|
PARD6B
|
par-6 partitioning defective 6 homolog beta (C. elegans)
|
|
chr2_-_72228427
|
2.099
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr10_+_88506320
|
2.076
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr1_-_48235148
|
1.930
|
NM_001194986
|
LOC388630
|
UPF0632 protein A
|
|
chr20_-_1113116
|
1.920
|
NM_018354
|
C20orf46
|
chromosome 20 open reading frame 46
|
|
chr16_-_87535106
|
1.880
|
NM_175931
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr22_+_24290738
|
1.766
|
NM_005160
|
ADRBK2
|
adrenergic, beta, receptor kinase 2
|
|
chr12_+_56230110
|
1.590
|
NM_004984
|
KIF5A
|
kinesin family member 5A
|
|
chr5_+_140187746
|
1.561
|
NM_018909
NM_031848
NM_031849
|
PCDHA6
|
protocadherin alpha 6
|
|
chr5_+_140325930
|
1.543
|
NM_018899
|
PCDHAC2
|
protocadherin alpha subfamily C, 2
|
|
chr11_-_13474101
|
1.500
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr2_-_219633481
|
1.450
|
NM_002181
|
IHH
|
Indian hedgehog
|
|
chr1_+_65385799
|
1.442
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr5_+_140160966
|
1.374
|
NM_018906
NM_031497
|
PCDHA3
|
protocadherin alpha 3
|
|
chr5_+_140215778
|
1.361
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr5_+_140235114
|
1.338
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr14_-_87863003
|
1.329
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr5_+_140166842
|
1.292
|
NM_018907
NM_031500
|
PCDHA4
|
protocadherin alpha 4
|
|
chr12_+_56435042
|
1.278
|
NM_138396
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9
|
|
chr15_-_27650218
|
1.241
|
NM_015307
|
FAM189A1
|
family with sequence similarity 189, member A1
|
|
chr12_-_23993830
|
1.241
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr22_+_38770610
|
1.228
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr5_+_140194152
|
1.222
|
NM_018910
NM_031852
|
PCDHA7
|
protocadherin alpha 7
|
|
chr12_-_55168168
|
1.208
|
NM_013267
|
GLS2
|
glutaminase 2 (liver, mitochondrial)
|
|
chr6_-_94185780
|
1.199
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr16_-_49742008
|
1.172
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr16_+_49139694
|
1.167
|
NM_033119
|
NKD1
|
naked cuticle homolog 1 (Drosophila)
|
|
chr9_-_10602722
|
1.157
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr4_-_77163609
|
1.149
|
NM_001565
|
CXCL10
|
chemokine (C-X-C motif) ligand 10
|
|
chr15_+_45263517
|
1.106
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr6_+_73388291
|
1.078
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr3_+_123385219
|
1.039
|
NM_000388
|
CASR
|
calcium-sensing receptor
|
|
chr5_+_140227951
|
1.015
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr18_+_74841262
|
1.012
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr12_-_7916686
|
1.006
|
NM_153449
|
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14
|
|
chr13_-_45859635
|
0.988
|
NM_025113
|
C13orf18
|
chromosome 13 open reading frame 18
|
|
chr12_-_108989882
|
0.986
|
NM_207435
|
C12orf76
|
chromosome 12 open reading frame 76
|
|
chr5_+_140286485
|
0.980
|
NM_018898
NM_031882
|
PCDHAC1
|
protocadherin alpha subfamily C, 1
|
|
chr19_-_16599986
|
0.969
|
NM_004831
|
MED26
|
mediator complex subunit 26
|
|
chr20_+_19141282
|
0.927
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr5_+_140181405
|
0.925
|
NM_018908
NM_031501
|
PCDHA5
|
protocadherin alpha 5
|
|
chr20_+_44083667
|
0.921
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr4_+_154083584
|
0.908
|
NM_033393
|
FHDC1
|
FH2 domain containing 1
|
|
chr1_+_78017894
|
0.900
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr9_-_111300350
|
0.898
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr4_-_41445693
|
0.893
|
NM_003924
|
PHOX2B
|
paired-like homeobox 2b
|
|
chr18_+_53253775
|
0.888
|
NM_004852
|
ONECUT2
|
one cut homeobox 2
|
|
chr6_-_97391779
|
0.880
|
NM_001143957
NM_030784
|
GPR63
|
G protein-coupled receptor 63
|
|
chr10_-_128884411
|
0.873
|
NM_001039762
|
FAM196A
|
family with sequence similarity 196, member A
|
|
chr17_+_38088097
|
0.857
|
NM_003632
|
CNTNAP1
|
contactin associated protein 1
|
|
chr22_+_38903825
|
0.851
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_-_205290921
|
0.823
|
NM_018566
|
YOD1
|
YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae)
|
|
chr16_-_49742651
|
0.817
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr7_-_15692798
|
0.806
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr4_-_78959532
|
0.804
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr5_-_148013908
|
0.796
|
NM_000870
|
HTR4
|
5-hydroxytryptamine (serotonin) receptor 4
|
|
chr1_-_85439315
|
0.778
|
NM_032184
|
SYDE2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans)
|
|
chr8_+_95722516
|
0.775
|
NM_001034915
NM_001122825
NM_001122826
NM_001122827
NM_017697
|
ESRP1
|
epithelial splicing regulatory protein 1
|
|
chr9_+_494661
|
0.774
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr18_-_44723116
|
0.771
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr12_-_101396459
|
0.760
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr5_+_166644420
|
0.741
|
NM_001122679
|
ODZ2
|
odz, odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr11_-_72530605
|
0.732
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr18_-_44729700
|
0.730
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr5_+_140200995
|
0.718
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr2_-_36384
|
0.715
|
NM_001077710
|
FAM110C
|
family with sequence similarity 110, member C
|
|
chr4_-_153675602
|
0.714
|
NM_033632
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr16_-_66827449
|
0.710
|
NM_024939
|
ESRP2
|
epithelial splicing regulatory protein 2
|
|
chr8_-_93176618
|
0.710
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr4_-_82612049
|
0.709
|
NM_152545
|
RASGEF1B
|
RasGEF domain family, member 1B
|
|
chr18_-_30057512
|
0.707
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr1_+_65386359
|
0.696
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chr3_-_53356573
|
0.670
|
NM_018403
|
DCP1A
|
DCP1 decapping enzyme homolog A (S. cerevisiae)
|
|
chr14_-_36058648
|
0.661
|
NM_003317
|
NKX2-1
|
NK2 homeobox 1
|
|
chr5_-_76970264
|
0.648
|
NM_032109
|
OTP
|
orthopedia homeobox
|
|
chr5_+_140207540
|
0.643
|
NM_014005
NM_031857
|
PCDHA9
|
protocadherin alpha 9
|
|
chr12_-_69600780
|
0.642
|
NM_002849
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr5_+_140146059
|
0.641
|
NM_018900
NM_031410
NM_031411
|
PCDHA1
|
protocadherin alpha 1
|
|
chr10_-_21503025
|
0.638
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr12_-_116283793
|
0.633
|
NM_000620
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr10_+_103103797
|
0.630
|
NM_003939
NM_033637
|
BTRC
|
beta-transducin repeat containing
|
|
chr2_-_98646367
|
0.628
|
NM_001160154
|
MGAT4A
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A
|
|
chr17_-_75427644
|
0.624
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr9_-_111122826
|
0.621
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr1_-_52604405
|
0.616
|
NM_032449
|
CC2D1B
|
coiled-coil and C2 domain containing 1B
|
|
chr11_-_44288188
|
0.614
|
NM_021926
|
ALX4
|
ALX homeobox 4
|
|
chr14_-_36059084
|
0.595
|
NM_001079668
|
NKX2-1
|
NK2 homeobox 1
|
|
chr12_-_38123027
|
0.593
|
NM_001173463
NM_001173464
NM_001173465
NM_017641
|
KIF21A
|
kinesin family member 21A
|
|
chr2_+_149349242
|
0.589
|
NM_004522
|
KIF5C
|
kinesin family member 5C
|
|
chr17_-_2250967
|
0.585
|
NM_020310
|
MNT
|
MAX binding protein
|
|
chr20_-_44713466
|
0.581
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr14_+_101297887
|
0.579
|
NM_001161725
NM_001161726
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr14_-_87859284
|
0.573
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr3_+_123385870
|
0.563
|
NM_001178065
|
CASR
|
calcium-sensing receptor
|
|
chr5_+_140154627
|
0.555
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr17_+_63252087
|
0.546
|
NM_004459
NM_182641
|
BPTF
|
bromodomain PHD finger transcription factor
|
|
chr17_+_32368611
|
0.544
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr17_+_56032271
|
0.542
|
NM_003620
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr8_-_93177057
|
0.541
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_+_231610438
|
0.541
|
NM_001144994
|
C2orf72
|
chromosome 2 open reading frame 72
|
|
chr18_-_59137488
|
0.540
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr12_-_61283474
|
0.536
|
NM_175895
|
C12orf61
|
chromosome 12 open reading frame 61
|
|
chr5_-_132976121
|
0.533
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr1_-_181654124
|
0.532
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr19_-_56215084
|
0.528
|
NM_002776
NM_001077500
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr14_-_98807574
|
0.519
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr4_+_76077229
|
0.519
|
NM_015393
|
PARM1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr18_-_30056431
|
0.504
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr17_+_16886491
|
0.485
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr8_+_100025806
|
0.485
|
NM_001142462
NM_053001
|
OSR2
|
odd-skipped related 2 (Drosophila)
|
|
chr10_+_104667991
|
0.483
|
NM_017649
NM_199076
NM_199077
|
CNNM2
|
cyclin M2
|
|
chr17_-_34015631
|
0.481
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr11_-_60685454
|
0.474
|
NM_017966
|
VPS37C
|
vacuolar protein sorting 37 homolog C (S. cerevisiae)
|
|
chr11_-_73787065
|
0.471
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr12_-_69434598
|
0.471
|
NM_130846
|
PTPRR
|
protein tyrosine phosphatase, receptor type, R
|
|
chr10_-_94040779
|
0.468
|
NM_014912
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr18_-_59185330
|
0.467
|
NM_002035
|
KDSR
|
3-ketodihydrosphingosine reductase
|
|
chr6_+_135544138
|
0.466
|
NM_001130172
NM_001130173
NM_001161656
NM_001161657
NM_001161658
NM_001161659
NM_001161660
NM_005375
|
MYB
|
v-myb myeloblastosis viral oncogene homolog (avian)
|
|
chr3_+_151609395
|
0.466
|
NM_014779
|
TSC22D2
|
TSC22 domain family, member 2
|
|
chr18_-_33399997
|
0.465
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr7_+_138566770
|
0.460
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr5_+_139473873
|
0.458
|
NM_005859
|
PURA
|
purine-rich element binding protein A
|
|
chr11_-_8571988
|
0.456
|
NM_030906
|
STK33
|
serine/threonine kinase 33
|
|
chr19_-_56214765
|
0.455
|
NM_145888
|
KLK10
|
kallikrein-related peptidase 10
|
|
chr2_-_121759237
|
0.451
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr3_-_129777618
|
0.448
|
NM_007354
|
C3orf27
|
chromosome 3 open reading frame 27
|
|
chr3_-_159306532
|
0.447
|
NM_001163678
NM_003030
NM_006884
|
SHOX2
|
short stature homeobox 2
|
|
chr16_-_87570716
|
0.444
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr12_-_101398503
|
0.438
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr18_+_53170718
|
0.437
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chrX_+_56275510
|
0.433
|
NM_007250
NM_001159296
|
KLF8
|
Kruppel-like factor 8
|
|
chr18_-_44731078
|
0.428
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_154436670
|
0.426
|
NM_052917
|
GALNT13
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 13 (GalNAc-T13)
|
|
chr20_+_44091219
|
0.426
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr1_+_226261374
|
0.419
|
NM_033131
|
WNT3A
|
wingless-type MMTV integration site family, member 3A
|
|
chr1_-_238842071
|
0.415
|
NM_022469
|
GREM2
|
gremlin 2
|
|
chr22_+_40702709
|
0.411
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr2_+_65308332
|
0.408
|
NM_001005386
NM_005722
|
ACTR2
|
ARP2 actin-related protein 2 homolog (yeast)
|
|
chr10_-_103525646
|
0.405
|
NM_006119
NM_033163
NM_033164
NM_033165
|
FGF8
|
fibroblast growth factor 8 (androgen-induced)
|
|
chr6_+_71179826
|
0.404
|
NM_001105531
NM_020819
|
FAM135A
|
family with sequence similarity 135, member A
|
|
chr1_-_50661716
|
0.402
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr5_+_170779264
|
0.397
|
NM_003862
|
FGF18
|
fibroblast growth factor 18
|
|
chr12_-_49708279
|
0.395
|
NM_001174125
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr7_+_140420500
|
0.395
|
NM_001195278
|
LOC100131199
|
transmembrane protein 178-like
|
|
chr1_-_149955415
|
0.387
|
NM_001172648
NM_001172649
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr8_-_4839670
|
0.381
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr13_-_20374895
|
0.378
|
NM_022459
|
XPO4
|
exportin 4
|
|
chr9_-_130574007
|
0.377
|
NM_006336
|
ZER1
|
zer-1 homolog (C. elegans)
|
|
chr1_-_149955870
|
0.376
|
NM_007185
|
CELF3
|
CUGBP, Elav-like family member 3
|
|
chr4_-_146320243
|
0.370
|
NM_001102653
|
OTUD4
|
OTU domain containing 4
|
|
chr6_-_134537699
|
0.370
|
NM_005627
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_118740069
|
0.369
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr4_-_68898294
|
0.369
|
NM_001031732
NM_133370
|
YTHDC1
|
YTH domain containing 1
|
|
chr4_+_72271218
|
0.364
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr2_+_61098200
|
0.364
|
NM_002618
|
PEX13
|
peroxisomal biogenesis factor 13
|
|
chr9_+_70584739
|
0.363
|
NM_138333
|
FAM122A
|
family with sequence similarity 122A
|
|
chr10_+_103882776
|
0.361
|
NM_015062
|
PPRC1
|
peroxisome proliferator-activated receptor gamma, coactivator-related 1
|
|
chr11_+_45899747
|
0.359
|
NM_152312
|
GYLTL1B
|
glycosyltransferase-like 1B
|
|
chr12_-_55686466
|
0.358
|
NM_014830
|
ZBTB39
|
zinc finger and BTB domain containing 39
|
|
chr19_-_55706156
|
0.355
|
NM_138334
|
JOSD2
|
Josephin domain containing 2
|
|
chr3_-_180272291
|
0.349
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr4_+_167013859
|
0.347
|
NM_012464
|
TLL1
|
tolloid-like 1
|
|
chr12_-_7979896
|
0.338
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr17_-_24645289
|
0.336
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr1_-_158136521
|
0.331
|
NM_012337
|
CCDC19
|
coiled-coil domain containing 19
|
|
chr1_+_179719338
|
0.331
|
NM_000721
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr2_+_151974849
|
0.330
|
NM_001177665
|
RIF1
|
RAP1 interacting factor homolog (yeast)
|
|
chr2_-_220116062
|
0.329
|
NM_001195731
|
CHPF
|
chondroitin polymerizing factor
|
|
chr14_-_19999475
|
0.326
|
NM_001100814
NM_144568
|
TMEM55B
|
transmembrane protein 55B
|
|
chr1_+_160734218
|
0.325
|
NM_144624
NM_175866
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr3_-_131095061
|
0.324
|
NM_001128224
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr12_-_106679038
|
0.322
|
NM_012406
|
PRDM4
|
PR domain containing 4
|
|
chr19_-_45016587
|
0.314
|
NM_004714
NM_006483
NM_006484
|
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B
|
|
chr19_-_14177980
|
0.311
|
NM_001008701
NM_014921
|
LPHN1
|
latrophilin 1
|
|
chr14_+_101345878
|
0.306
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr5_-_95323353
|
0.304
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr18_+_27926566
|
0.301
|
NM_001191324
NM_198128
|
RNF138
|
ring finger protein 138
|
|
chr5_+_140241976
|
0.300
|
NM_018904
NM_031865
|
PCDHA13
|
protocadherin alpha 13
|
|
chr15_-_91433165
|
0.299
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr17_+_22645232
|
0.298
|
NM_015626
NM_134265
|
WSB1
|
WD repeat and SOCS box containing 1
|
|
chr3_+_11009419
|
0.297
|
NM_003042
|
SLC6A1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr13_+_99056887
|
0.291
|
NM_206808
|
CLYBL
|
citrate lyase beta like
|
|
chr14_-_38971370
|
0.289
|
NM_203301
|
FBXO33
|
F-box protein 33
|
|
chr7_+_113842284
|
0.288
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr10_-_93992982
|
0.286
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr3_+_134775946
|
0.284
|
NM_001134423
|
CDV3
|
CDV3 homolog (mouse)
|
|
chr12_-_23628774
|
0.279
|
NM_178010
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr14_-_30746431
|
0.279
|
NM_015382
|
HECTD1
|
HECT domain containing 1
|
|
chr11_+_9552809
|
0.278
|
NM_001143976
|
WEE1
|
WEE1 homolog (S. pombe)
|
|
chr4_-_153493133
|
0.277
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr11_-_1725924
|
0.274
|
NM_001170820
|
LOC402778
|
CD225 family protein FLJ76511
|
|
chr22_-_49413326
|
0.272
|
NM_000487
NM_001085425
NM_001085426
NM_001085427
NM_001085428
|
ARSA
|
arylsulfatase A
|
|
chr16_-_23068010
|
0.271
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr12_-_52960168
|
0.269
|
NM_012117
|
CBX5
|
chromobox homolog 5
|
|
chrX_-_139414890
|
0.266
|
NM_005634
|
SOX3
|
SRY (sex determining region Y)-box 3
|
|
chr17_-_62671767
|
0.263
|
NM_014877
|
HELZ
|
helicase with zinc finger
|
|
chr18_-_43711452
|
0.262
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|