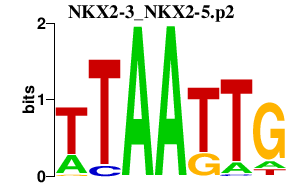
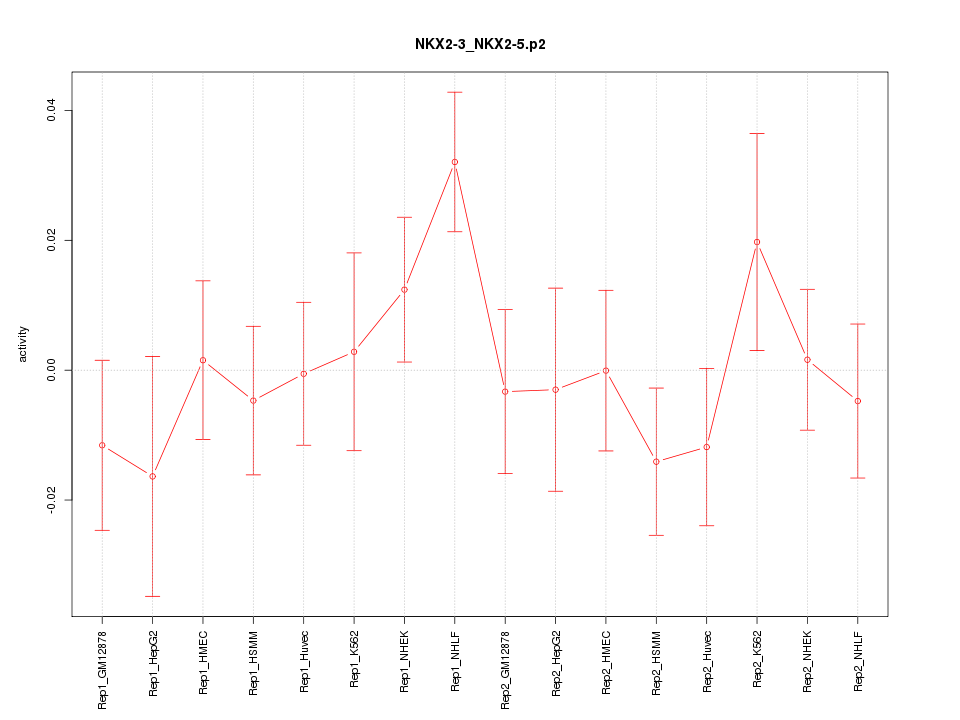
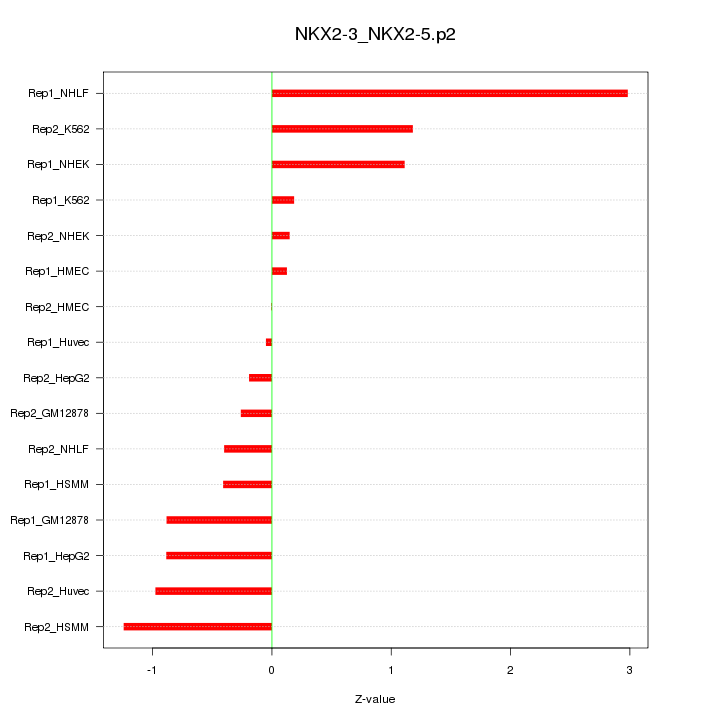
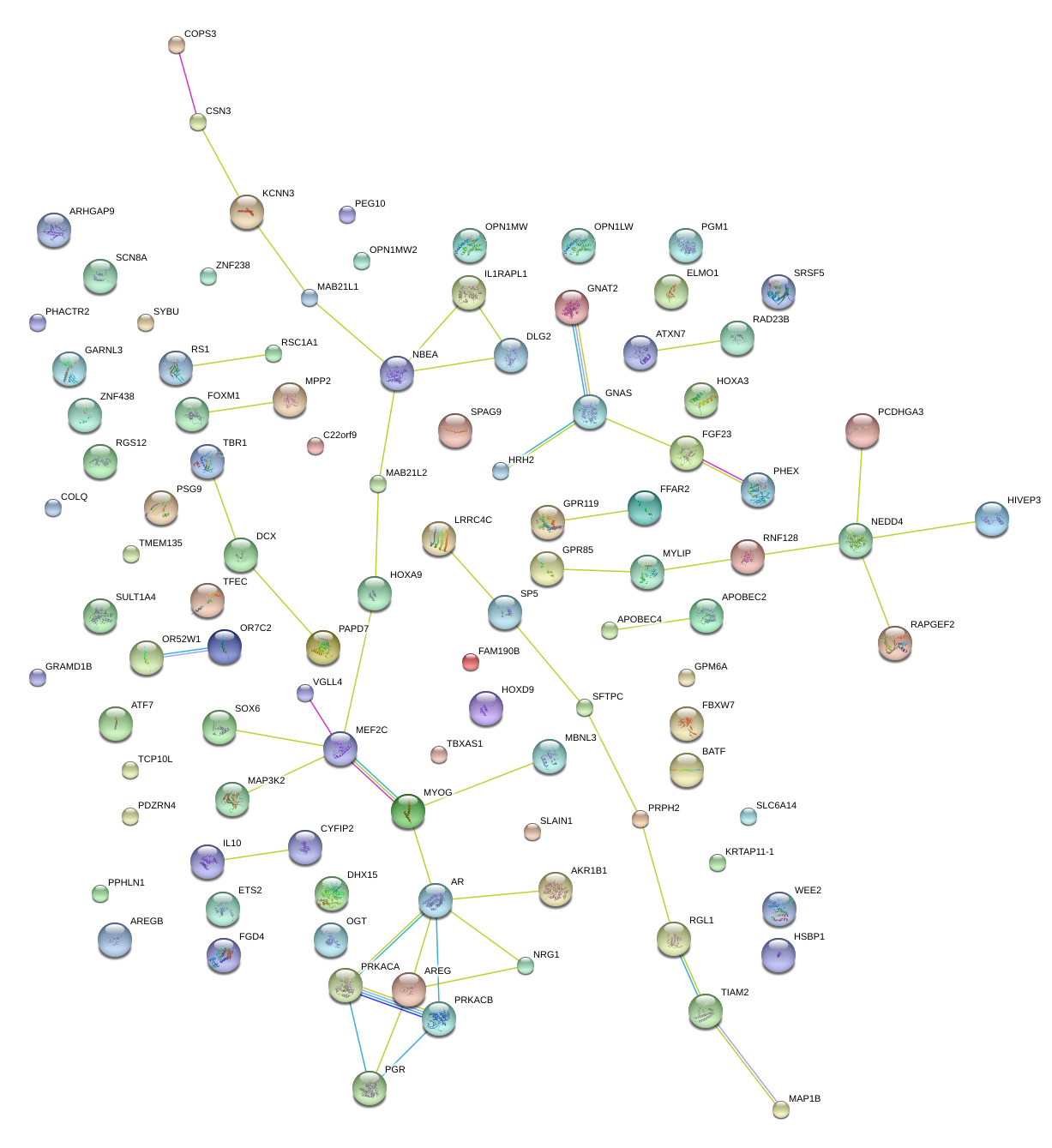
|
chr7_-_27171673
|
2.156
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171648
|
1.921
|
|
HOXA9
|
homeobox A9
|
|
chrX_+_70682204
|
1.815
|
|
OGT
|
O-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase)
|
|
chr7_-_27171600
|
1.809
|
|
HOXA9
|
homeobox A9
|
|
chr2_+_37282516
|
1.441
|
|
LOC100505876
|
hypothetical LOC100505876
|
|
chr5_+_71538456
|
1.320
|
|
MAP1B
|
microtubule-associated protein 1B
|
|
chrX_+_21960841
|
1.259
|
NM_000444
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked
|
|
chrX_-_129347101
|
1.020
|
NM_178471
|
GPR119
|
G protein-coupled receptor 119
|
|
chr7_+_94135384
|
0.990
|
|
PEG10
|
paternally expressed 10
|
|
chrX_+_66705407
|
0.946
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr6_+_155579788
|
0.866
|
NM_001010927
|
TIAM2
|
T-cell lymphoma invasion and metastasis 2
|
|
chr20_+_56919233
|
0.848
|
|
GNAS
|
GNAS complex locus
|
|
chr10_-_31328426
|
0.846
|
NM_001143769
|
ZNF438
|
zinc finger protein 438
|
|
chr19_+_40632456
|
0.794
|
NM_005306
|
FFAR2
|
free fatty acid receptor 2
|
|
chr5_+_140772691
|
0.792
|
NM_018913
NM_032090
|
PCDHGA10
|
protocadherin gamma subfamily A, 10
|
|
chrX_+_28515479
|
0.767
|
NM_014271
|
IL1RAPL1
|
interleukin 1 receptor accessory protein-like 1
|
|
chr9_+_129066576
|
0.746
|
NM_032293
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3
|
|
chr17_-_46479127
|
0.738
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr5_+_156628929
|
0.721
|
NM_014376
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr12_-_4358968
|
0.699
|
NM_020638
|
FGF23
|
fibroblast growth factor 23
|
|
chr3_-_15538164
|
0.685
|
NM_005677
NM_080539
|
COLQ
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase
|
|
chr1_-_109957195
|
0.677
|
NM_005272
|
GNAT2
|
guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 2
|
|
chr7_+_139175420
|
0.674
|
NM_001061
NM_001166253
NM_030984
|
TBXAS1
|
thromboxane A synthase 1 (platelet)
|
|
chrX_-_110541029
|
0.661
|
NM_000555
|
DCX
|
doublecortin
|
|
chr12_+_50342814
|
0.652
|
NM_001177984
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr1_+_63861474
|
0.649
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr13_+_77213295
|
0.643
|
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr12_-_52281050
|
0.636
|
NM_001130059
|
ATF7
|
activating transcription factor 7
|
|
chr3_-_11598829
|
0.636
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr4_-_153493133
|
0.629
|
NM_018315
|
FBXW7
|
F-box and WD repeat domain containing 7
|
|
chr11_+_122936246
|
0.627
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chr11_-_40272239
|
0.625
|
NM_020929
|
LRRC4C
|
leucine rich repeat containing 4C
|
|
chr11_+_86710076
|
0.616
|
|
TMEM135
|
transmembrane protein 135
|
|
chr11_-_16454493
|
0.609
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr1_-_153109349
|
0.607
|
NM_002249
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr1_-_205012348
|
0.589
|
NM_000572
|
IL10
|
interleukin 10
|
|
chr7_-_112513379
|
0.584
|
NM_001146266
NM_001146267
|
GPR85
|
G protein-coupled receptor 85
|
|
chr22_-_16127520
|
0.573
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3
|
|
chr4_+_71142893
|
0.559
|
NM_005212
|
CSN3
|
casein kappa
|
|
chr9_+_109131814
|
0.555
|
|
RAD23B
|
RAD23 homolog B (S. cerevisiae)
|
|
chr5_-_88155360
|
0.554
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr4_+_160408447
|
0.545
|
NM_014247
|
RAPGEF2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr1_+_84402962
|
0.540
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chrX_-_131401319
|
0.537
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chrX_+_111005934
|
0.532
|
NM_001195576
NM_001195578
|
LOC100329135
|
hypothetical LOC100329135
|
|
chr7_-_36991189
|
0.528
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_+_69307914
|
0.525
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr12_+_40117751
|
0.524
|
NM_013377
|
PDZRN4
|
PDZ domain containing ring finger 4
|
|
chrX_+_115481774
|
0.520
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr21_-_32879617
|
0.520
|
NM_144659
|
TCP10L
|
t-complex 10 (mouse)-like
|
|
chr7_-_27133163
|
0.517
|
NM_153631
|
HOXA3
|
homeobox A3
|
|
chr5_+_140703784
|
0.506
|
NM_018916
NM_032011
|
PCDHGA3
|
protocadherin gamma subfamily A, 3
|
|
chr13_-_34948758
|
0.504
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr6_+_16251260
|
0.503
|
|
MYLIP
|
myosin regulatory light chain interacting protein
|
|
chr2_-_127817143
|
0.493
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr1_-_201321999
|
0.488
|
NM_002479
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr7_-_115458047
|
0.487
|
|
TFEC
|
transcription factor EC
|
|
chr5_+_175041069
|
0.486
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr2_+_171280046
|
0.484
|
NM_001003845
|
SP5
|
Sp5 transcription factor
|
|
chr13_+_34948977
|
0.482
|
|
NBEA
|
neurobeachin
|
|
chrX_+_105823723
|
0.480
|
NM_024539
|
RNF128
|
ring finger protein 128
|
|
chr3_+_63859114
|
0.468
|
NM_001177387
|
ATXN7
|
ataxin 7
|
|
chr6_+_41128917
|
0.465
|
NM_006789
|
APOBEC2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2
|
|
chrX_+_153101278
|
0.455
|
NM_000513
NM_001048181
|
OPN1MW
OPN1MW2
|
opsin 1 (cone pigments), medium-wave-sensitive
opsin 1 (cone pigments), medium-wave-sensitive 2
|
|
chr6_+_143971009
|
0.445
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr11_+_6176951
|
0.445
|
NM_001005178
|
OR52W1
|
olfactory receptor, family 52, subfamily W, member 1
|
|
chr1_-_181889070
|
0.444
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr1_+_242281175
|
0.435
|
NM_205768
|
ZNF238
|
zinc finger protein 238
|
|
chr7_-_115458010
|
0.434
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chr21_+_39117064
|
0.429
|
|
ETS2
|
v-ets erythroblastosis virus E26 oncogene homolog 2 (avian)
|
|
chr1_+_556968
|
0.426
|
|
|
|
|
chr1_-_42156913
|
0.424
|
|
|
|
|
chr12_+_32546243
|
0.417
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr21_-_31175722
|
0.416
|
NM_175858
|
KRTAP11-1
|
keratin associated protein 11-1
|
|
chr1_-_42156957
|
0.415
|
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chr22_-_43987010
|
0.410
|
NM_015264
|
KIAA0930
|
KIAA0930
|
|
chr14_+_75058518
|
0.410
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr11_-_83071084
|
0.409
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_+_182040837
|
0.407
|
|
RGL1
|
ral guanine nucleotide dissociation stimulator-like 1
|
|
chr5_+_140762551
|
0.406
|
NM_018921
NM_032089
|
PCDHGA9
|
protocadherin gamma subfamily A, 9
|
|
chr4_+_3341521
|
0.403
|
NM_198227
|
RGS12
|
regulator of G-protein signaling 12
|
|
chr8_-_110689396
|
0.401
|
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_-_88235624
|
0.401
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr2_+_161981046
|
0.399
|
|
TBR1
|
T-box, brain, 1
|
|
chr10_+_86174665
|
0.398
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr7_+_141054621
|
0.396
|
NM_001105558
|
WEE2
|
WEE1 homolog 2 (S. pombe)
|
|
chr4_-_176970895
|
0.392
|
|
GPM6A
|
glycoprotein M6A
|
|
chr15_-_53996597
|
0.391
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chrX_-_18600108
|
0.385
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr6_+_29187565
|
0.382
|
NM_001005216
|
OR2J3
|
olfactory receptor, family 2, subfamily J, member 3
|
|
chrX_+_102852492
|
0.374
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr10_+_126620681
|
0.373
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr2_+_17861266
|
0.371
|
NM_001105569
|
MSGN1
|
mesogenin 1
|
|
chr2_+_171280182
|
0.370
|
|
|
|
|
chr1_+_242281199
|
0.369
|
|
ZNF238
|
zinc finger protein 238
|
|
chr1_+_84540921
|
0.369
|
NM_001134664
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr1_+_166516901
|
0.367
|
NM_005149
|
TBX19
|
T-box 19
|
|
chr15_+_97990705
|
0.366
|
NM_001130928
|
MEF2A
|
myocyte enhancer factor 2A
|
|
chr22_-_36836620
|
0.365
|
NM_025045
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr13_+_34948885
|
0.363
|
|
NBEA
|
neurobeachin
|
|
chr15_-_59308770
|
0.357
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_46894239
|
0.355
|
NM_000316
|
PTH1R
|
parathyroid hormone 1 receptor
|
|
chr5_+_140848905
|
0.355
|
NM_018929
NM_032407
|
PCDHGC5
|
protocadherin gamma subfamily C, 5
|
|
chr8_+_42671718
|
0.355
|
NM_000749
|
CHRNB3
|
cholinergic receptor, nicotinic, beta 3
|
|
chr2_+_161981138
|
0.354
|
|
TBR1
|
T-box, brain, 1
|
|
chr18_+_18989786
|
0.347
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr3_+_14835472
|
0.347
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr11_+_33306652
|
0.346
|
|
HIPK3
|
homeodomain interacting protein kinase 3
|
|
chr19_+_4442610
|
0.345
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr15_-_59308708
|
0.344
|
|
RORA
|
RAR-related orphan receptor A
|
|
chr19_-_3723208
|
0.341
|
NM_032753
|
RAX2
|
retina and anterior neural fold homeobox 2
|
|
chr1_+_17817397
|
0.341
|
|
ARHGEF10L
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr12_-_6103945
|
0.339
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr1_+_221168388
|
0.336
|
NM_032890
|
DISP1
|
dispatched homolog 1 (Drosophila)
|
|
chr16_+_7322751
|
0.333
|
NM_145891
NM_145892
NM_145893
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr10_+_69539255
|
0.330
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr1_-_166149867
|
0.328
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr6_-_114490701
|
0.327
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr12_-_51173238
|
0.326
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr14_+_99633570
|
0.323
|
|
EVL
|
Enah/Vasp-like
|
|
chr21_-_36774257
|
0.323
|
NM_001146079
NM_144492
|
CLDN14
|
claudin 14
|
|
chr4_-_186970366
|
0.322
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr8_-_40874485
|
0.319
|
NM_001135731
NM_024645
|
ZMAT4
|
zinc finger, matrin-type 4
|
|
chr3_-_57209319
|
0.318
|
NM_003865
|
HESX1
|
HESX homeobox 1
|
|
chr7_-_96471544
|
0.318
|
|
DLX6-AS1
|
DLX6 antisense RNA 1 (non-protein coding)
|
|
chr10_+_86121526
|
0.317
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr13_+_112746908
|
0.317
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr22_-_36836478
|
0.316
|
|
BAIAP2L2
|
BAI1-associated protein 2-like 2
|
|
chr1_+_35623688
|
0.316
|
|
ZMYM4
|
zinc finger, MYM-type 4
|
|
chr14_+_75058591
|
0.314
|
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr1_-_201321711
|
0.314
|
|
MYOG
|
myogenin (myogenic factor 4)
|
|
chr17_+_24394281
|
0.313
|
|
PIPOX
|
pipecolic acid oxidase
|
|
chr3_-_11585264
|
0.313
|
NM_001128221
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chrX_-_10495476
|
0.311
|
NM_001193278
NM_001193279
NM_001193280
NM_001193281
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr13_+_97856148
|
0.309
|
|
|
|
|
chrX_-_21686079
|
0.307
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr19_-_46504831
|
0.306
|
|
|
|
|
chr4_+_2853434
|
0.303
|
|
ADD1
|
adducin 1 (alpha)
|
|
chr2_+_48649662
|
0.300
|
NM_172311
|
STON1-GTF2A1L
|
STON1-GTF2A1L readthrough
|
|
chr2_+_161980865
|
0.298
|
NM_006593
|
TBR1
|
T-box, brain, 1
|
|
chr1_-_154041393
|
0.292
|
|
GON4L
|
gon-4-like (C. elegans)
|
|
chr10_-_75071205
|
0.291
|
|
MYOZ1
|
myozenin 1
|
|
chrX_+_44588192
|
0.291
|
NM_022076
|
DUSP21
|
dual specificity phosphatase 21
|
|
chr7_+_128802719
|
0.291
|
NM_001130722
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr7_+_147918589
|
0.290
|
NM_145304
|
C7orf33
|
chromosome 7 open reading frame 33
|
|
chr22_+_21006814
|
0.289
|
|
|
|
|
chr1_-_42157077
|
0.289
|
NM_001127714
NM_024503
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3
|
|
chrX_-_133620103
|
0.289
|
NM_021796
|
PLAC1
|
placenta-specific 1
|
|
chr21_-_30836051
|
0.288
|
NM_181612
|
KRTAP19-6
|
keratin associated protein 19-6
|
|
chr1_-_32157845
|
0.288
|
NM_001195100
NM_001195101
|
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2
|
|
chr12_-_16650697
|
0.288
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr7_-_26198673
|
0.288
|
|
HNRNPA2B1
|
heterogeneous nuclear ribonucleoprotein A2/B1
|
|
chr7_+_129771865
|
0.285
|
NM_001127441
NM_001127442
NM_080385
|
CPA5
|
carboxypeptidase A5
|
|
chr3_+_16191159
|
0.285
|
NM_054110
|
GALNTL2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 2
|
|
chrX_-_102829621
|
0.284
|
NM_001142419
NM_001142420
NM_001142421
NM_001142422
NM_001142427
NM_001142428
NM_001142431
NM_001142432
|
MORF4L2
|
mortality factor 4 like 2
|
|
chrX_+_100111352
|
0.283
|
NM_001012990
NM_001162490
NM_001162491
|
ARL13A
|
ADP-ribosylation factor-like 13A
|
|
chrX_-_128616594
|
0.282
|
NM_017413
|
APLN
|
apelin
|
|
chr4_-_176971175
|
0.280
|
NM_201591
|
GPM6A
|
glycoprotein M6A
|
|
chr12_-_121767272
|
0.279
|
NM_006018
|
GPR109B
|
G protein-coupled receptor 109B
|
|
chr2_+_28892366
|
0.278
|
NM_001008779
|
SPDYA
|
speedy homolog A (Xenopus laevis)
|
|
chrX_+_70438352
|
0.277
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr7_+_104539548
|
0.276
|
|
MLL5
|
myeloid/lymphoid or mixed-lineage leukemia 5 (trithorax homolog, Drosophila)
|
|
chr4_+_88939725
|
0.276
|
NM_004967
|
IBSP
|
integrin-binding sialoprotein
|
|
chr6_+_136214495
|
0.276
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr8_-_7275061
|
0.275
|
NM_018661
|
DEFB103B
|
defensin, beta 103B
|
|
chr6_+_12824873
|
0.273
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr4_-_40631397
|
0.273
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr4_-_2200755
|
0.273
|
NM_181808
|
POLN
|
polymerase (DNA directed) nu
|
|
chr11_+_122936145
|
0.272
|
|
GRAMD1B
|
GRAM domain containing 1B
|
|
chrX_+_68752635
|
0.272
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr12_+_100512877
|
0.270
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chrX_+_144716619
|
0.269
|
NM_004709
|
CXorf1
|
chromosome X open reading frame 1
|
|
chrX_-_137649180
|
0.265
|
NM_033642
|
FGF13
|
fibroblast growth factor 13
|
|
chrX_+_107174855
|
0.265
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr17_-_36209672
|
0.265
|
NM_181535
|
KRT28
|
keratin 28
|
|
chr2_-_55090759
|
0.264
|
NM_007008
|
RTN4
|
reticulon 4
|
|
chr5_-_168204430
|
0.264
|
|
|
|
|
chr5_+_176494690
|
0.263
|
|
NSD1
|
nuclear receptor binding SET domain protein 1
|
|
chrX_-_114158388
|
0.263
|
NM_000640
|
IL13RA2
|
interleukin 13 receptor, alpha 2
|
|
chr10_-_64246129
|
0.261
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr15_-_68781662
|
0.261
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr3_+_130730171
|
0.260
|
NM_000539
|
RHO
|
rhodopsin
|
|
chrX_-_13247946
|
0.258
|
|
ATXN3L
|
ataxin 3-like
|
|
chrX_-_43717864
|
0.254
|
NM_000266
|
NDP
|
Norrie disease (pseudoglioma)
|
|
chr10_-_98326435
|
0.252
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr10_+_124660206
|
0.252
|
NM_001029888
|
FAM24A
|
family with sequence similarity 24, member A
|
|
chr5_+_176744037
|
0.252
|
NM_001167579
NM_003052
|
SLC34A1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr17_-_36347197
|
0.251
|
NM_015515
|
KRT23
|
keratin 23 (histone deacetylase inducible)
|
|
chr2_-_210726539
|
0.250
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr13_-_102517196
|
0.250
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr20_+_43784401
|
0.249
|
NM_178455
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4
|
|
chr11_-_57275828
|
0.249
|
NM_001145101
|
BTBD18
|
BTB (POZ) domain containing 18
|
|
chr11_+_4799126
|
0.248
|
NM_001004753
|
OR51F2
|
olfactory receptor, family 51, subfamily F, member 2
|
|
chr1_+_15862887
|
0.248
|
|
|
|
|
chr12_+_54700955
|
0.243
|
NM_022465
|
IKZF4
|
IKAROS family zinc finger 4 (Eos)
|
|
chr10_-_75071498
|
0.242
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chr3_+_39484073
|
0.240
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr2_-_80384454
|
0.236
|
|
LRRTM1
|
leucine rich repeat transmembrane neuronal 1
|
|
chr4_+_94969064
|
0.235
|
NM_005172
|
ATOH1
|
atonal homolog 1 (Drosophila)
|
|
chr8_+_67096344
|
0.235
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|