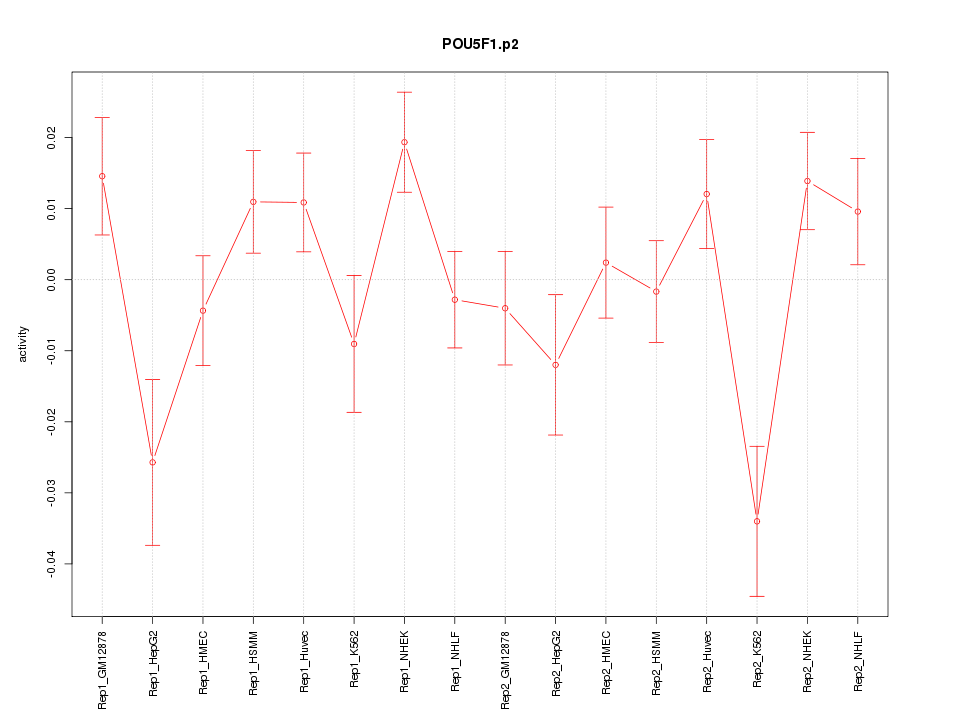
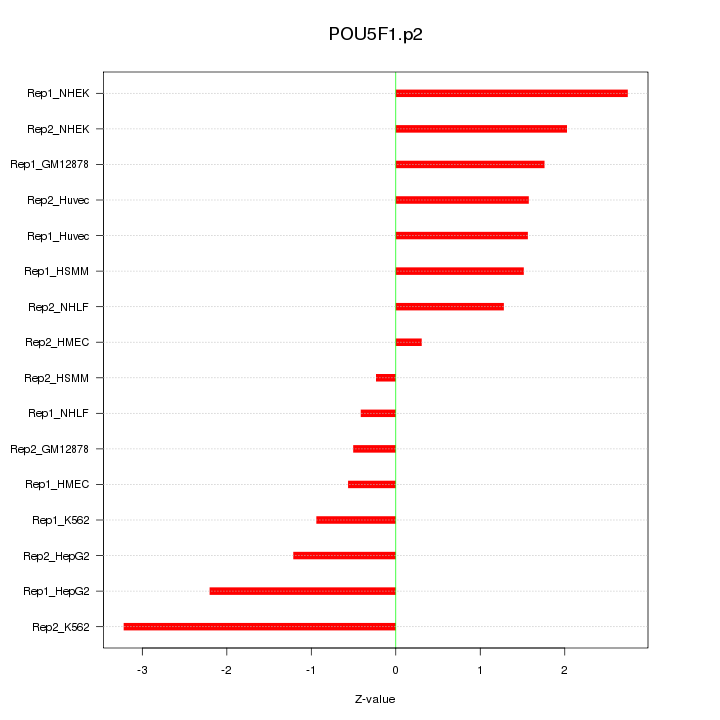
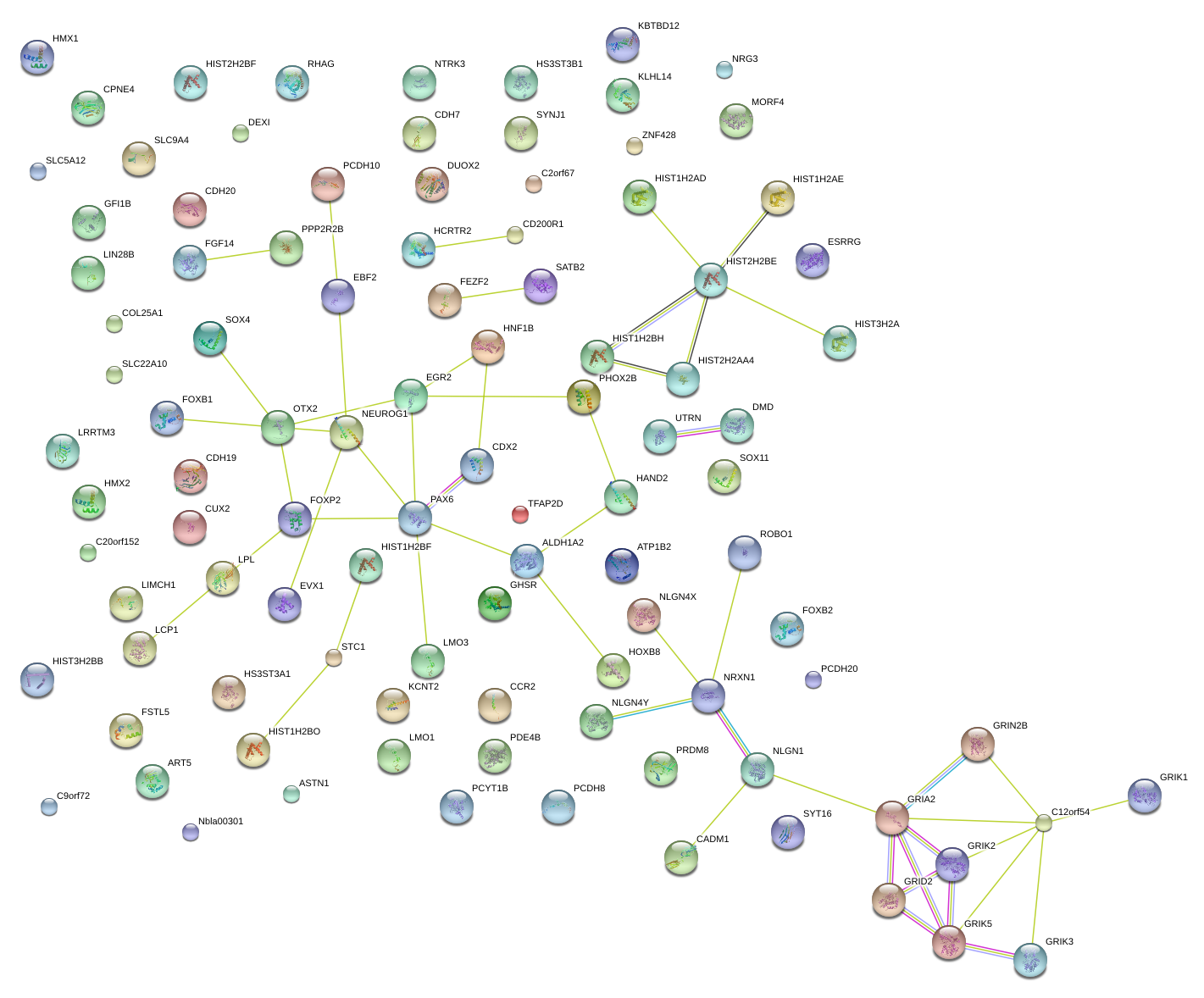
|
chr12_-_16649211
|
4.105
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr4_-_110443247
|
3.354
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr10_+_68355797
|
2.738
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr2_+_5750229
|
2.646
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr15_+_26832300
|
2.569
|
|
LOC100289656
|
Dexi homolog (mouse) pseudogene
|
|
chr17_-_33178987
|
2.534
|
|
HNF1B
|
HNF1 homeobox B
|
|
chr13_-_52320774
|
2.500
|
NM_002590
NM_032949
|
PCDH8
|
protocadherin 8
|
|
chrX_-_6156655
|
2.461
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr11_-_3620060
|
2.457
|
NM_053017
NM_001079536
|
ART5
|
ADP-ribosyltransferase 5
|
|
chr13_-_101852028
|
2.411
|
NM_175929
|
FGF14
|
fibroblast growth factor 14
|
|
chr13_-_60887210
|
2.307
|
|
PCDH20
|
protocadherin 20
|
|
chr17_+_7494964
|
2.303
|
NM_001678
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr17_+_7495140
|
2.213
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr17_-_33179118
|
2.095
|
NM_000458
NM_001165923
|
HNF1B
|
HNF1 homeobox B
|
|
chr4_+_158361185
|
2.059
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr4_-_41445479
|
2.025
|
|
PHOX2B
|
paired-like homeobox 2b
|
|
chrX_-_33267646
|
2.022
|
NM_000109
|
DMD
|
dystrophin
|
|
chr4_+_41395493
|
1.932
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_+_105511615
|
1.905
|
NM_001004317
|
LIN28B
|
lin-28 homolog B (C. elegans)
|
|
chr15_-_86600658
|
1.809
|
NM_001007156
NM_001012338
NM_002530
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3
|
|
chr1_-_214963279
|
1.798
|
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_+_124897627
|
1.692
|
NM_005519
|
HMX2
|
H6 family homeobox 2
|
|
chr1_-_215329569
|
1.685
|
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr17_-_44047299
|
1.671
|
NM_024016
|
HOXB8
|
homeobox B8
|
|
chr11_-_31789360
|
1.651
|
|
PAX6
|
paired box 6
|
|
chr1_-_147665783
|
1.591
|
|
HIST2H2BF
|
histone cluster 2, H2bf
|
|
chr6_+_50789215
|
1.586
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr8_-_25958556
|
1.572
|
|
EBF2
|
early B-cell factor 2
|
|
chr1_-_175400573
|
1.555
|
NM_004319
NM_207108
|
ASTN1
|
astrotactin 1
|
|
chr3_+_174598937
|
1.468
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr11_-_31789334
|
1.460
|
|
PAX6
|
paired box 6
|
|
chr17_+_7495436
|
1.457
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr3_+_129124591
|
1.457
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr12_-_16650697
|
1.438
|
NM_018640
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_+_61532293
|
1.430
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr14_-_56342097
|
1.425
|
NM_172337
|
OTX2
|
orthodenticle homeobox 2
|
|
chrX_-_24575146
|
1.384
|
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr11_-_31789431
|
1.382
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr15_+_58083737
|
1.373
|
|
|
|
|
chr7_+_27248688
|
1.355
|
NM_001989
|
EVX1
|
even-skipped homeobox 1
|
|
chr6_-_49712483
|
1.351
|
NM_000324
|
RHAG
|
Rh-associated glycoprotein
|
|
chr1_-_194844061
|
1.348
|
NM_198503
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr4_-_174774368
|
1.324
|
NM_006792
|
MORF4
|
mortality factor 4
|
|
chr8_-_23768242
|
1.313
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr3_+_46370228
|
1.302
|
NM_001123041
NM_001123396
|
CCR2
|
chemokine (C-C motif) receptor 2
|
|
chr17_+_14145051
|
1.299
|
NM_006041
|
HS3ST3B1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1
|
|
chr15_+_58083712
|
1.295
|
NM_012182
|
FOXB1
|
forkhead box B1
|
|
chr10_-_64248932
|
1.285
|
NM_001136178
|
EGR2
|
early growth response 2
|
|
chr12_-_14024187
|
1.281
|
NM_000834
|
GRIN2B
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2B
|
|
chr1_-_226712140
|
1.279
|
NM_033445
|
HIST3H2A
|
histone cluster 3, H2a
|
|
chr3_-_62334229
|
1.278
|
NM_018008
|
FEZF2
|
FEZ family zinc finger 2
|
|
chr5_-_146237826
|
1.274
|
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr11_+_62813947
|
1.267
|
NM_001039752
|
SLC22A10
|
solute carrier family 22, member 10
|
|
chr1_+_226712430
|
1.265
|
NM_175055
|
HIST3H2BB
|
histone cluster 3, H2bb
|
|
chr12_+_109956210
|
1.262
|
NM_015267
|
CUX2
|
cut-like homeobox 2
|
|
chr11_-_114880275
|
1.260
|
NM_001098517
NM_014333
|
CADM1
|
cell adhesion molecule 1
|
|
chr4_+_174688183
|
1.237
|
|
NBLA00301
|
Nbla00301
|
|
chr20_+_34019942
|
1.231
|
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr19_-_48815572
|
1.223
|
|
ZNF428
|
zinc finger protein 428
|
|
chr5_-_134899537
|
1.221
|
NM_006161
|
NEUROG1
|
neurogenin 1
|
|
chr10_+_83625049
|
1.220
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr2_-_50428395
|
1.217
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr15_-_56145140
|
1.209
|
NM_003888
NM_170696
|
ALDH1A2
|
aldehyde dehydrogenase 1 family, member A2
|
|
chr18_-_28606839
|
1.196
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr11_-_26700121
|
1.195
|
NM_178498
|
SLC5A12
|
solute carrier family 5 (sodium/glucose cotransporter), member 12
|
|
chr9_+_134843918
|
1.174
|
NM_001135031
NM_004188
|
GFI1B
|
growth factor independent 1B transcription repressor
|
|
chr4_-_163304567
|
1.159
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chr4_+_134289893
|
1.146
|
NM_020815
NM_032961
|
PCDH10
|
protocadherin 10
|
|
chr2_-_200038055
|
1.145
|
NM_001172517
|
SATB2
|
SATB homeobox 2
|
|
chr3_-_79899748
|
1.139
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr15_-_43193614
|
1.136
|
|
DUOX2
|
dual oxidase 2
|
|
chr3_-_173648896
|
1.132
|
NM_004122
NM_198407
|
GHSR
|
growth hormone secretagogue receptor
|
|
chr15_-_43193637
|
1.124
|
NM_014080
|
DUOX2
|
dual oxidase 2
|
|
chr4_-_174686918
|
1.107
|
|
HAND2
|
heart and neural crest derivatives expressed 2
|
|
chr18_+_57308754
|
1.106
|
NM_031891
|
CDH20
|
cadherin 20, type 2
|
|
chr13_-_27441423
|
1.097
|
|
CDX2
|
caudal type homeobox 2
|
|
chr6_+_55147029
|
1.089
|
NM_001526
|
HCRTR2
|
hypocretin (orexin) receptor 2
|
|
chr4_-_41579325
|
1.087
|
|
|
|
|
chr7_+_113513828
|
1.083
|
|
FOXP2
|
forkhead box P2
|
|
chr13_-_27441290
|
1.076
|
NM_001265
|
CDX2
|
caudal type homeobox 2
|
|
chr5_-_146238336
|
1.074
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr4_+_93444572
|
1.070
|
NM_001510
|
GRID2
|
glutamate receptor, ionotropic, delta 2
|
|
chrX_-_24575375
|
1.063
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr1_+_66230977
|
1.051
|
NM_001037340
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr3_-_133236447
|
1.046
|
NM_130808
|
CPNE4
|
copine IV
|
|
chr2_+_102456193
|
1.045
|
NM_001011552
|
SLC9A4
|
solute carrier family 9 (sodium/hydrogen exchanger), member 4
|
|
chr6_+_101953581
|
1.040
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr8_+_19841209
|
1.030
|
|
LPL
|
lipoprotein lipase
|
|
chr11_-_31789168
|
1.030
|
|
PAX6
|
paired box 6
|
|
chr5_-_88004891
|
1.028
|
|
LOC645323
|
hypothetical LOC645323
|
|
chr2_-_200033445
|
0.993
|
|
SATB2
|
SATB homeobox 2
|
|
chr4_+_81325447
|
0.988
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr15_+_58083798
|
0.987
|
|
FOXB1
|
forkhead box B1
|
|
chr2_-_210726539
|
0.986
|
|
C2orf67
|
chromosome 2 open reading frame 67
|
|
chr18_+_61568467
|
0.980
|
NM_033646
|
CDH7
|
cadherin 7, type 2
|
|
chrX_+_9391314
|
0.971
|
NM_001139466
NM_001139467
NM_001139468
|
TBL1X
|
transducin (beta)-like 1X-linked
|
|
chr20_+_17155669
|
0.960
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr5_+_149526533
|
0.958
|
NM_001804
|
CDX1
|
caudal type homeobox 1
|
|
chr3_-_24511180
|
0.957
|
NM_001128176
NM_000461
NM_001128177
|
THRB
|
thyroid hormone receptor, beta (erythroblastic leukemia viral (v-erb-a) oncogene homolog 2, avian)
|
|
chr8_+_19841120
|
0.943
|
|
LPL
|
lipoprotein lipase
|
|
chr20_+_17155680
|
0.941
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr6_+_32003380
|
0.936
|
|
C2
|
complement component 2
|
|
chr11_+_119616112
|
0.935
|
NM_014352
|
POU2F3
|
POU class 2 homeobox 3
|
|
chrX_-_109570113
|
0.929
|
NM_001171689
|
AMMECR1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1
|
|
chr1_+_168381811
|
0.927
|
NM_001136107
|
METTL11B
|
methyltransferase like 11B
|
|
chr3_+_182912769
|
0.925
|
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr7_+_113513726
|
0.916
|
|
FOXP2
|
forkhead box P2
|
|
chr1_-_175400665
|
0.914
|
|
ASTN1
|
astrotactin 1
|
|
chr13_-_27441143
|
0.912
|
|
CDX2
|
caudal type homeobox 2
|
|
chr22_+_29848923
|
0.912
|
NM_001002837
|
INPP5J
|
inositol polyphosphate-5-phosphatase J
|
|
chr1_-_175400441
|
0.911
|
|
ASTN1
|
astrotactin 1
|
|
chr8_+_19841010
|
0.905
|
|
LPL
|
lipoprotein lipase
|
|
chr12_+_54231249
|
0.900
|
NM_001005494
|
OR6C4
|
olfactory receptor, family 6, subfamily C, member 4
|
|
chrX_+_99726445
|
0.899
|
NM_022144
|
TNMD
|
tenomodulin
|
|
chr12_-_80676562
|
0.898
|
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr3_+_182912405
|
0.891
|
NM_003106
|
SOX2
|
SRY (sex determining region Y)-box 2
|
|
chr13_+_57103789
|
0.891
|
NM_001040429
|
PCDH17
|
protocadherin 17
|
|
chr10_-_97190885
|
0.886
|
NM_001034954
NM_001034955
NM_001034956
NM_001034957
NM_006434
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr12_-_10846435
|
0.886
|
NM_023919
|
TAS2R7
|
taste receptor, type 2, member 7
|
|
chr10_+_50488352
|
0.879
|
NM_003055
|
SLC18A3
|
solute carrier family 18 (vesicular acetylcholine), member 3
|
|
chr4_+_2013517
|
0.873
|
NM_001168243
NM_001141936
|
C4orf48
|
chromosome 4 open reading frame 48
|
|
chr3_-_195601283
|
0.870
|
NM_004488
|
GP5
|
glycoprotein V (platelet)
|
|
chr8_-_21701616
|
0.866
|
|
GFRA2
|
GDNF family receptor alpha 2
|
|
chr7_+_94377145
|
0.863
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr18_-_29412148
|
0.856
|
|
|
|
|
chr4_-_154929657
|
0.852
|
NM_003013
|
SFRP2
|
secreted frizzled-related protein 2
|
|
chr7_-_50103335
|
0.847
|
NM_001159878
NM_007009
|
ZPBP
|
zona pellucida binding protein
|
|
chr18_+_59374372
|
0.844
|
NM_080474
|
SERPINB12
|
serpin peptidase inhibitor, clade B (ovalbumin), member 12
|
|
chr3_-_152173475
|
0.843
|
NM_001195794
NM_174878
|
CLRN1
|
clarin 1
|
|
chr5_-_147142249
|
0.839
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr15_-_40537002
|
0.839
|
NM_022473
|
ZFP106
|
zinc finger protein 106 homolog (mouse)
|
|
chr12_-_16650687
|
0.838
|
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr12_+_52713098
|
0.837
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr3_-_193928038
|
0.830
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr6_-_128281434
|
0.829
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr6_+_108593907
|
0.828
|
NM_003269
|
NR2E1
|
nuclear receptor subfamily 2, group E, member 1
|
|
chr5_-_111340421
|
0.826
|
NM_001142474
NM_001142475
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr20_+_17155617
|
0.825
|
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr3_-_166397142
|
0.824
|
NM_014926
|
SLITRK3
|
SLIT and NTRK-like family, member 3
|
|
chr8_-_93181091
|
0.823
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_192767494
|
0.819
|
|
TMEFF2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chr7_-_76667045
|
0.819
|
NM_006682
|
FGL2
|
fibrinogen-like 2
|
|
chr12_-_94469357
|
0.818
|
NM_032147
|
USP44
|
ubiquitin specific peptidase 44
|
|
chr7_+_45899621
|
0.804
|
|
|
|
|
chr8_+_19840861
|
0.802
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr1_-_151296611
|
0.800
|
NM_005988
|
SPRR2A
|
small proline-rich protein 2A
|
|
chr10_-_64246129
|
0.798
|
NM_000399
NM_001136177
NM_001136179
|
EGR2
|
early growth response 2
|
|
chr18_+_29412538
|
0.798
|
NM_030632
|
ASXL3
|
additional sex combs like 3 (Drosophila)
|
|
chr10_+_95838801
|
0.786
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr20_+_9442995
|
0.784
|
|
C20orf103
|
chromosome 20 open reading frame 103
|
|
chr12_+_56290229
|
0.783
|
NM_001111270
|
ARHGEF25
|
Rho guanine nucleotide exchange factor (GEF) 25
|
|
chr12_-_16652202
|
0.781
|
NM_001001395
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr2_-_238813284
|
0.764
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr16_+_23754772
|
0.761
|
NM_002738
NM_212535
|
PRKCB
|
protein kinase C, beta
|
|
chr14_-_33489708
|
0.758
|
NM_022073
|
EGLN3
|
egl nine homolog 3 (C. elegans)
|
|
chr3_+_135996990
|
0.758
|
|
EPHB1
|
EPH receptor B1
|
|
chr3_+_143824955
|
0.757
|
NM_002670
|
PLS1
|
plastin 1
|
|
chr12_-_10853940
|
0.756
|
NM_023917
|
TAS2R9
|
taste receptor, type 2, member 9
|
|
chr16_+_23754822
|
0.756
|
|
PRKCB
|
protein kinase C, beta
|
|
chr3_-_140879463
|
0.749
|
NM_178177
|
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3
|
|
chr13_-_34948758
|
0.740
|
NM_005584
|
MAB21L1
|
mab-21-like 1 (C. elegans)
|
|
chr4_-_164754106
|
0.737
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr13_-_60887655
|
0.737
|
NM_022843
|
PCDH20
|
protocadherin 20
|
|
chr10_-_64246080
|
0.737
|
|
EGR2
|
early growth response 2
|
|
chr1_-_194844299
|
0.735
|
|
KCNT2
|
potassium channel, subfamily T, member 2
|
|
chr14_-_105623358
|
0.735
|
|
IGHA1
IGHD
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant delta
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr5_-_177950161
|
0.734
|
NM_173465
|
COL23A1
|
collagen, type XXIII, alpha 1
|
|
chr7_-_36991189
|
0.733
|
NM_001039459
|
ELMO1
|
engulfment and cell motility 1
|
|
chr14_+_64077171
|
0.726
|
|
HSPA2
|
heat shock 70kDa protein 2
|
|
chr6_+_132901119
|
0.720
|
NM_175057
|
TAAR9
|
trace amine associated receptor 9 (gene/pseudogene)
|
|
chr22_-_43784444
|
0.719
|
NM_001135862
|
PHF21B
|
PHD finger protein 21B
|
|
chr17_-_72045218
|
0.716
|
NM_134268
|
CYGB
|
cytoglobin
|
|
chr20_-_49852353
|
0.716
|
NM_020436
|
SALL4
|
sal-like 4 (Drosophila)
|
|
chr5_+_140767953
|
0.715
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr7_-_141320041
|
0.712
|
NM_176817
|
TAS2R38
|
taste receptor, type 2, member 38
|
|
chr4_+_41309690
|
0.711
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr19_+_40511909
|
0.709
|
NM_001185099
NM_001185100
NM_001185101
NM_001771
|
CD22
|
CD22 molecule
|
|
chr2_+_165860528
|
0.697
|
NM_001040143
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr8_-_95289945
|
0.695
|
NM_004063
|
CDH17
|
cadherin 17, LI cadherin (liver-intestine)
|
|
chr4_-_100575313
|
0.692
|
NM_001166504
NM_000673
|
ADH7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr17_-_75427644
|
0.688
|
NM_003655
|
CBX4
|
chromobox homolog 4
|
|
chr8_+_104900591
|
0.683
|
NM_014677
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr4_+_41309482
|
0.677
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr18_+_61569136
|
0.676
|
NM_004361
|
CDH7
|
cadherin 7, type 2
|
|
chr2_-_104834344
|
0.667
|
|
LOC100506421
|
hypothetical LOC100506421
|
|
chr7_+_28441758
|
0.661
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr7_-_92693716
|
0.658
|
NM_001039372
|
HEPACAM2
|
HEPACAM family member 2
|
|
chr12_-_61832856
|
0.653
|
NM_000706
|
AVPR1A
|
arginine vasopressin receptor 1A
|
|
chr6_-_26151863
|
0.653
|
NM_021062
|
HIST1H2BB
|
histone cluster 1, H2bb
|
|
chr17_-_34635446
|
0.652
|
NM_198993
|
STAC2
|
SH3 and cysteine rich domain 2
|
|
chr11_-_57171554
|
0.651
|
|
YPEL4
|
yippee-like 4 (Drosophila)
|
|
chr3_+_1109619
|
0.648
|
NM_014461
|
CNTN6
|
contactin 6
|
|
chr16_+_23754860
|
0.647
|
|
PRKCB
|
protein kinase C, beta
|
|
chr2_-_89294466
|
0.646
|
|
IGKC
|
immunoglobulin kappa constant
|
|
chr4_-_47534815
|
0.646
|
NM_006587
|
CORIN
|
corin, serine peptidase
|
|
chr1_-_151280201
|
0.645
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr4_+_41309668
|
0.643
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr11_-_84312085
|
0.643
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr7_-_31347032
|
0.642
|
NM_022728
|
NEUROD6
|
neurogenic differentiation 6
|
|
chr8_+_75899326
|
0.642
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|