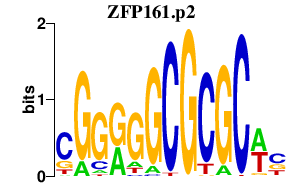
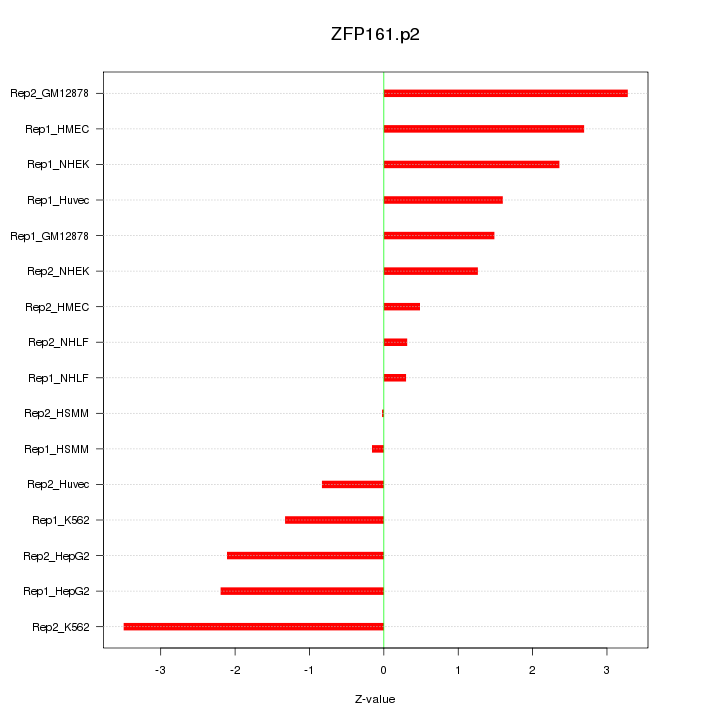
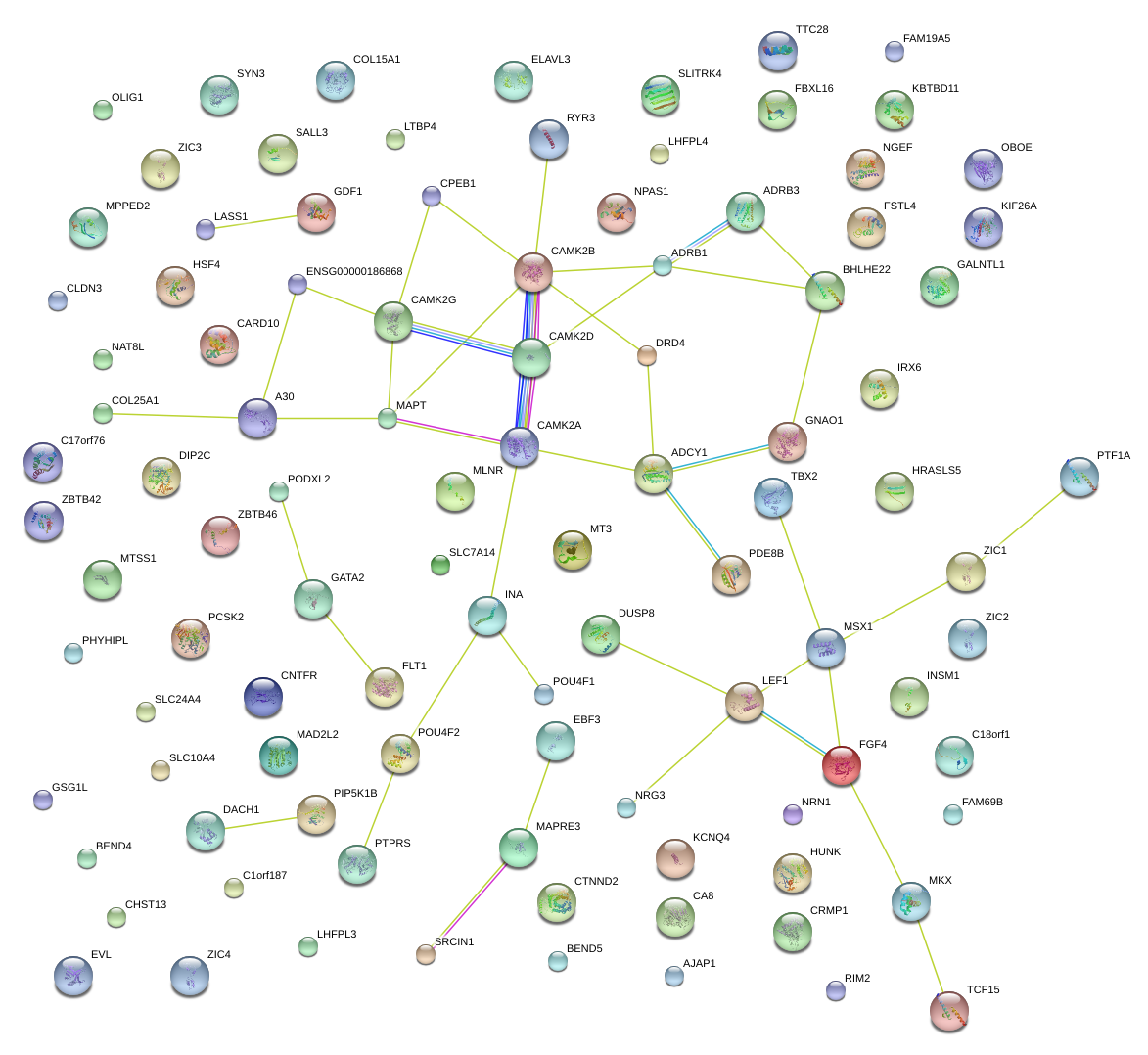
|
chr9_+_138726737
|
4.246
|
NM_152421
|
FAM69B
|
family with sequence similarity 69, member B
|
|
chr8_+_1937711
|
3.566
|
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr11_-_1549719
|
3.263
|
NM_004420
|
DUSP8
|
dual specificity phosphatase 8
|
|
chr2_-_233501069
|
2.983
|
NM_001114090
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr22_-_27405708
|
2.873
|
NM_001145418
|
TTC28
|
tetratricopeptide repeat domain 28
|
|
chr3_+_128830656
|
2.628
|
NM_015720
|
PODXL2
|
podocalyxin-like 2
|
|
chr20_+_20296744
|
2.271
|
NM_002196
|
INSM1
|
insulinoma-associated 1
|
|
chr13_-_78075683
|
2.248
|
NM_006237
|
POU4F1
|
POU class 4 homeobox 1
|
|
chr10_-_725522
|
2.223
|
NM_014974
|
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila)
|
|
chr10_+_105027431
|
1.963
|
|
INA
|
internexin neuronal intermediate filament protein, alpha
|
|
chr20_-_538909
|
1.926
|
NM_004609
|
TCF15
|
transcription factor 15 (basic helix-loop-helix)
|
|
chr4_-_5945596
|
1.880
|
NM_001014809
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr20_-_61933012
|
1.859
|
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr3_-_129689394
|
1.848
|
NM_001145662
|
GATA2
|
GATA binding protein 2
|
|
chr10_+_23521464
|
1.801
|
NM_178161
|
PTF1A
|
pancreas specific transcription factor, 1a
|
|
chr16_-_27982277
|
1.782
|
NM_001109763
|
GSG1L
|
GSG1-like
|
|
chr4_+_2031036
|
1.762
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr22_-_37569679
|
1.755
|
|
|
|
|
chr19_-_11452439
|
1.724
|
NM_001420
NM_032281
|
ELAVL3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr13_+_99432258
|
1.675
|
NM_007129
|
ZIC2
|
Zic family member 2 (odd-paired homolog, Drosophila)
|
|
chr1_-_11674264
|
1.654
|
NM_001127325
|
MAD2L2
|
MAD2 mitotic arrest deficient-like 2 (yeast)
|
|
chr20_+_17155617
|
1.650
|
NM_002594
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr4_-_110443247
|
1.648
|
NM_032518
NM_198721
|
COL25A1
|
collagen, type XXV, alpha 1
|
|
chr21_+_33364420
|
1.648
|
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr17_+_56832255
|
1.641
|
|
TBX2
|
T-box 2
|
|
chr16_+_55180767
|
1.628
|
NM_005954
|
MT3
|
metallothionein 3
|
|
chr16_+_53915971
|
1.627
|
NM_024335
|
IRX6
|
iroquois homeobox 6
|
|
chr8_+_65656164
|
1.584
|
|
BHLHE22
|
basic helix-loop-helix family, member e22
|
|
chr1_+_41022066
|
1.544
|
NM_004700
NM_172163
|
KCNQ4
|
potassium voltage-gated channel, KQT-like subfamily, member 4
|
|
chr11_-_30564338
|
1.535
|
NM_001145399
|
MPPED2
|
metallophosphoesterase domain containing 2
|
|
chr3_+_127743868
|
1.518
|
|
CHST13
|
carbohydrate (chondroitin 4) sulfotransferase 13
|
|
chr21_+_33364316
|
1.513
|
NM_138983
|
OLIG1
|
oligodendrocyte transcription factor 1
|
|
chr20_+_17155669
|
1.511
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr11_+_627268
|
1.501
|
NM_000797
|
DRD4
|
dopamine receptor D4
|
|
chr16_-_695719
|
1.485
|
NM_153350
|
FBXL16
|
F-box and leucine-rich repeat protein 16
|
|
chr17_-_34015631
|
1.479
|
NM_025248
|
SRCIN1
|
SRC kinase signaling inhibitor 1
|
|
chr9_-_34579679
|
1.476
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr18_-_21184824
|
1.474
|
|
|
|
|
chr2_+_104837004
|
1.469
|
|
|
|
|
chr22_+_47263919
|
1.465
|
NM_001082967
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5
|
|
chr5_-_132976121
|
1.450
|
NM_015082
|
FSTL4
|
follistatin-like 4
|
|
chr14_+_104338423
|
1.444
|
|
ZBTB42
|
zinc finger and BTB domain containing 42
|
|
chr18_+_74841262
|
1.441
|
NM_171999
|
SALL3
|
sal-like 3 (Drosophila)
|
|
chr14_+_91859289
|
1.425
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr16_+_54782802
|
1.420
|
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr19_+_45799060
|
1.415
|
NM_001042545
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr8_-_125809831
|
1.415
|
NM_014751
|
MTSS1
|
metastasis suppressor 1
|
|
chr4_-_109308893
|
1.407
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr17_+_41327808
|
1.404
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr3_-_9570412
|
1.404
|
NM_198560
|
LHFPL4
|
lipoma HMGIC fusion partner-like 4
|
|
chr17_+_56831951
|
1.384
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr5_-_132976094
|
1.383
|
|
FSTL4
|
follistatin-like 4
|
|
chr10_+_115793795
|
1.379
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr7_-_44331544
|
1.374
|
NM_001220
NM_172078
NM_172079
NM_172080
NM_172081
NM_172082
NM_172083
NM_172084
|
CAMK2B
|
calcium/calmodulin-dependent protein kinase II beta
|
|
chr19_-_5291700
|
1.343
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr14_+_68796621
|
1.334
|
|
GALNTL1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1
|
|
chr7_-_72822471
|
1.327
|
NM_001306
|
CLDN3
|
claudin 3
|
|
chr19_-_18867868
|
1.323
|
NM_021267
NM_198207
NM_001492
|
LASS1
GDF1
|
LAG1 homolog, ceramide synthase 1
growth differentiation factor 1
|
|
chr6_-_5952631
|
1.303
|
NM_016588
|
NRN1
|
neuritin 1
|
|
chr16_+_65754788
|
1.295
|
NM_001040667
NM_001538
|
HSF4
|
heat shock transcription factor 4
|
|
chr13_-_27967215
|
1.295
|
NM_001159920
NM_001160030
NM_001160031
NM_002019
|
FLT1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor)
|
|
chr7_+_45580704
|
1.285
|
|
ADCY1
|
adenylate cyclase 1 (brain)
|
|
chr15_+_31390454
|
1.278
|
NM_001036
|
RYR3
|
ryanodine receptor 3
|
|
chr19_+_52215982
|
1.270
|
NM_002517
|
NPAS1
|
neuronal PAS domain protein 1
|
|
chr1_+_4614964
|
1.263
|
NM_001042478
NM_018836
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr10_+_60606352
|
1.260
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr13_-_71338144
|
1.248
|
|
DACH1
|
dachshund homolog 1 (Drosophila)
|
|
chr13_+_48692472
|
1.239
|
NM_001507
|
MLNR
|
motilin receptor
|
|
chr10_-_28074752
|
1.237
|
NM_173576
|
MKX
|
mohawk homeobox
|
|
chr11_-_69299351
|
1.235
|
NM_002007
|
FGF4
|
fibroblast growth factor 4
|
|
chr4_+_48180033
|
1.226
|
NM_152679
|
SLC10A4
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 4
|
|
chr15_-_81113676
|
1.218
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr22_-_36245155
|
1.217
|
NM_014550
|
CARD10
|
caspase recruitment domain family, member 10
|
|
chr17_+_56832492
|
1.212
|
|
TBX2
|
T-box 2
|
|
chrX_-_142549984
|
1.206
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr10_-_131652007
|
1.204
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr11_-_63014986
|
1.191
|
NM_001146728
NM_001146729
NM_054108
|
HRASLS5
|
HRAS-like suppressor family, member 5
|
|
chr21_+_32167175
|
1.183
|
NM_014586
|
HUNK
|
hormonally up-regulated Neu-associated kinase
|
|
chr1_+_11674365
|
1.172
|
NM_198545
|
C1orf187
|
chromosome 1 open reading frame 187
|
|
chr18_-_29412148
|
1.171
|
|
|
|
|
chr8_-_61356436
|
1.168
|
NM_004056
|
CA8
|
carbonic anhydrase VIII
|
|
chr3_-_129694717
|
1.159
|
NM_032638
|
GATA2
|
GATA binding protein 2
|
|
chr1_-_49014993
|
1.159
|
NM_024603
|
BEND5
|
BEN domain containing 5
|
|
chr1_-_1525309
|
1.157
|
|
LOC643988
|
hypothetical LOC643988
|
|
chr22_-_31784328
|
1.152
|
NM_001135774
|
SYN3
|
synapsin III
|
|
chr8_+_104582151
|
1.149
|
NM_001100117
|
RIMS2
|
regulating synaptic membrane exocytosis 2
|
|
chr17_-_16336204
|
1.142
|
NM_001113567
NM_207387
|
C17orf76
|
chromosome 17 open reading frame 76
|
|
chr3_-_171786468
|
1.135
|
NM_020949
|
SLC7A14
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 14
|
|
chr14_+_103674812
|
1.130
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr4_-_41848874
|
1.127
|
|
BEND4
|
BEN domain containing 4
|
|
chr17_+_41327662
|
1.126
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr4_+_147779492
|
1.117
|
NM_004575
|
POU4F2
|
POU class 4 homeobox 2
|
|
chr20_+_17155680
|
1.115
|
|
PCSK2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr5_-_11956964
|
1.114
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr9_+_100745531
|
1.113
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr9_+_70509925
|
1.109
|
NM_003558
|
PIP5K1B
|
phosphatidylinositol-4-phosphate 5-kinase, type I, beta
|
|
chr5_+_76542461
|
1.107
|
NM_001029851
NM_001029852
NM_001029853
NM_001029854
NM_003719
|
PDE8B
|
phosphodiesterase 8B
|
|
chr8_-_37941970
|
1.106
|
|
ADRB3
|
adrenergic, beta-3-, receptor
|
|
chr4_+_4912272
|
1.104
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr18_+_13208777
|
1.095
|
NM_181481
NM_181482
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr14_+_99507848
|
1.093
|
|
EVL
|
Enah/Vasp-like
|
|
chr17_+_41327449
|
1.087
|
NM_001123066
NM_001123067
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr10_+_83625049
|
1.083
|
NM_001010848
NM_001165972
|
NRG3
|
neuregulin 3
|
|
chr2_+_171280944
|
1.066
|
|
SP5
|
Sp5 transcription factor
|
|
chr1_-_215377718
|
1.063
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_+_94823221
|
1.063
|
NM_057157
|
CYP26A1
|
cytochrome P450, family 26, subfamily A, polypeptide 1
|
|
chr11_-_6396864
|
1.057
|
NM_001164
|
APBB1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65)
|
|
chr2_+_112372671
|
1.035
|
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr16_+_25610847
|
1.033
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr9_+_17568952
|
1.031
|
NM_003026
|
SH3GL2
|
SH3-domain GRB2-like 2
|
|
chr2_-_119632788
|
1.031
|
NM_182528
|
C1QL2
|
complement component 1, q subcomponent-like 2
|
|
chr4_-_41849333
|
1.027
|
|
BEND4
|
BEN domain containing 4
|
|
chr16_+_54782751
|
1.024
|
NM_020988
NM_138736
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O
|
|
chr5_+_75734759
|
1.018
|
NM_006633
|
IQGAP2
|
IQ motif containing GTPase activating protein 2
|
|
chr12_+_50271283
|
1.015
|
NM_014191
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit
|
|
chr10_-_131652364
|
1.004
|
|
EBF3
|
early B-cell factor 3
|
|
chr13_-_26232777
|
1.000
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr11_-_2248757
|
0.992
|
NM_005170
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr12_+_2032649
|
0.978
|
NM_000719
NM_001129827
NM_001129829
NM_001129830
NM_001129831
NM_001129832
NM_001129833
NM_001129834
NM_001129835
NM_001129836
NM_001129837
NM_001129838
NM_001129839
NM_001129840
NM_001129841
NM_001129842
NM_001129843
NM_001129844
NM_001129846
NM_001167623
NM_001167624
NM_001167625
NM_199460
|
CACNA1C
|
calcium channel, voltage-dependent, L type, alpha 1C subunit
|
|
chr2_-_42573980
|
0.976
|
|
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3
|
|
chr17_+_4434582
|
0.972
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr14_-_62581656
|
0.971
|
NM_139318
NM_172375
|
KCNH5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5
|
|
chr19_+_38979502
|
0.966
|
NM_001129994
NM_001129995
NM_024076
|
KCTD15
|
potassium channel tetramerisation domain containing 15
|
|
chr1_-_21850874
|
0.962
|
NM_001145658
|
RAP1GAP
|
RAP1 GTPase activating protein
|
|
chrX_-_34585287
|
0.960
|
NM_031442
|
TMEM47
|
transmembrane protein 47
|
|
chr3_-_127558747
|
0.956
|
NM_014079
|
KLF15
|
Kruppel-like factor 15
|
|
chr10_-_60606195
|
0.955
|
|
|
|
|
chr4_-_109307325
|
0.954
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr12_+_93066370
|
0.950
|
NM_005761
|
PLXNC1
|
plexin C1
|
|
chr4_+_19864332
|
0.950
|
NM_004787
|
SLIT2
|
slit homolog 2 (Drosophila)
|
|
chrX_-_51256166
|
0.949
|
NM_018159
|
NUDT11
|
nudix (nucleoside diphosphate linked moiety X)-type motif 11
|
|
chr19_-_18868395
|
0.946
|
|
LASS1
|
LAG1 homolog, ceramide synthase 1
|
|
chr2_-_39040981
|
0.945
|
|
LOC375196
|
hypothetical LOC375196
|
|
chr19_+_47509274
|
0.943
|
NM_173633
|
TMEM145
|
transmembrane protein 145
|
|
chr11_-_33847834
|
0.938
|
|
LMO2
|
LIM domain only 2 (rhombotin-like 1)
|
|
chr14_-_77034853
|
0.935
|
NM_182509
NM_199296
|
ISM2
|
isthmin 2 homolog (zebrafish)
|
|
chr17_+_41327792
|
0.935
|
|
MAPT
|
microtubule-associated protein tau
|
|
chr15_+_97462675
|
0.928
|
NM_015286
NM_145728
|
SYNM
|
synemin, intermediate filament protein
|
|
chr14_+_73128162
|
0.927
|
NM_152331
|
ACOT4
|
acyl-CoA thioesterase 4
|
|
chr8_-_125809559
|
0.920
|
|
MTSS1
|
metastasis suppressor 1
|
|
chr15_-_75711743
|
0.919
|
NM_032808
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr22_+_48951286
|
0.910
|
NM_001160300
NM_052839
|
PANX2
|
pannexin 2
|
|
chr8_-_133561738
|
0.905
|
|
KCNQ3
|
potassium voltage-gated channel, KQT-like subfamily, member 3
|
|
chr3_+_129690735
|
0.904
|
|
|
|
|
chr3_-_135097111
|
0.901
|
NM_016577
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr14_+_91859904
|
0.900
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr8_-_144583718
|
0.896
|
NM_201589
|
MAFA
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian)
|
|
chr22_+_42137955
|
0.894
|
NM_001044370
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr8_+_1909437
|
0.889
|
NM_014867
|
KBTBD11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr19_-_7845325
|
0.887
|
NM_001190467
|
FLJ22184
|
hypothetical protein FLJ22184
|
|
chr4_+_72271218
|
0.882
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr19_+_40325993
|
0.880
|
NM_022006
|
FXYD7
|
FXYD domain containing ion transport regulator 7
|
|
chr17_+_8865547
|
0.878
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr10_+_24023680
|
0.878
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr1_+_224803226
|
0.874
|
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr4_+_158361185
|
0.872
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr11_-_64268126
|
0.872
|
NM_001098671
|
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated)
|
|
chr11_-_110088330
|
0.871
|
NM_020809
|
ARHGAP20
|
Rho GTPase activating protein 20
|
|
chr1_+_154878498
|
0.866
|
|
BCAN
|
brevican
|
|
chr2_-_238813284
|
0.862
|
|
HES6
|
hairy and enhancer of split 6 (Drosophila)
|
|
chr3_-_135096986
|
0.860
|
|
RAB6B
|
RAB6B, member RAS oncogene family
|
|
chr11_-_61441536
|
0.860
|
NM_013401
|
RAB3IL1
|
RAB3A interacting protein (rabin3)-like 1
|
|
chr14_+_69021149
|
0.858
|
NM_001161498
|
UPF0639
|
UPF0639 protein
|
|
chr16_+_1979959
|
0.856
|
NM_004209
|
SYNGR3
|
synaptogyrin 3
|
|
chrX_-_18282698
|
0.855
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr2_-_47650973
|
0.853
|
NM_022055
|
KCNK12
|
potassium channel, subfamily K, member 12
|
|
chr15_-_75711785
|
0.850
|
|
LINGO1
|
leucine rich repeat and Ig domain containing 1
|
|
chr12_+_48641545
|
0.848
|
NM_001651
|
AQP5
|
aquaporin 5
|
|
chr19_+_10392128
|
0.838
|
NM_001111307
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific
|
|
chr7_+_154943466
|
0.836
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr13_+_112671497
|
0.833
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr10_-_104169458
|
0.833
|
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr16_-_47873183
|
0.830
|
NM_004352
|
CBLN1
|
cerebellin 1 precursor
|
|
chr1_+_4614528
|
0.824
|
|
AJAP1
|
adherens junctions associated protein 1
|
|
chr11_-_2248374
|
0.821
|
|
ASCL2
|
achaete-scute complex homolog 2 (Drosophila)
|
|
chr2_+_112372651
|
0.814
|
NM_006343
|
MERTK
|
c-mer proto-oncogene tyrosine kinase
|
|
chr1_-_111018798
|
0.812
|
|
KCNA3
|
potassium voltage-gated channel, shaker-related subfamily, member 3
|
|
chr2_+_936534
|
0.810
|
NM_018968
|
SNTG2
|
syntrophin, gamma 2
|
|
chr10_+_122206455
|
0.808
|
NM_001030059
|
PPAPDC1A
|
phosphatidic acid phosphatase type 2 domain containing 1A
|
|
chr20_-_3097070
|
0.806
|
NM_014731
|
ProSAPiP1
|
ProSAPiP1 protein
|
|
chrX_+_38305634
|
0.802
|
NM_004615
|
TSPAN7
|
tetraspanin 7
|
|
chr1_+_224803123
|
0.800
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr6_+_107917965
|
0.795
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr18_-_70275441
|
0.794
|
NM_001044369
|
FAM69C
|
family with sequence similarity 69, member C
|
|
chr22_+_38183480
|
0.787
|
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase
|
|
chr5_-_83716346
|
0.781
|
NM_005711
|
EDIL3
|
EGF-like repeats and discoidin I-like domains 3
|
|
chrX_-_142550535
|
0.776
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr12_+_120944174
|
0.772
|
NM_001024808
NM_020993
|
BCL7A
|
B-cell CLL/lymphoma 7A
|
|
chr7_-_27180398
|
0.772
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr15_-_38362078
|
0.769
|
NM_001190479
|
LOC100131244
|
hypothetical protein LOC100131244
|
|
chr3_-_49882308
|
0.765
|
NM_024046
|
CAMKV
|
CaM kinase-like vesicle-associated
|
|
chr9_-_100598315
|
0.759
|
NM_173551
|
ANKS6
|
ankyrin repeat and sterile alpha motif domain containing 6
|
|
chr22_+_42138034
|
0.755
|
|
MPPED1
|
metallophosphoesterase domain containing 1
|
|
chr5_-_146238336
|
0.755
|
NM_004576
NM_181675
NM_001127381
|
PPP2R2B
|
protein phosphatase 2, regulatory subunit B, beta
|
|
chr7_+_113513828
|
0.754
|
|
FOXP2
|
forkhead box P2
|
|
chr1_-_202921103
|
0.754
|
NM_006338
NM_201630
|
LRRN2
|
leucine rich repeat neuronal 2
|
|
chr13_-_43259032
|
0.751
|
NM_017993
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1
|
|
chr1_+_66031443
|
0.750
|
NM_002600
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific
|
|
chr1_-_111019051
|
0.747
|
|
|
|
|
chr8_+_121892719
|
0.747
|
|
|
|