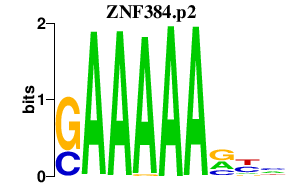
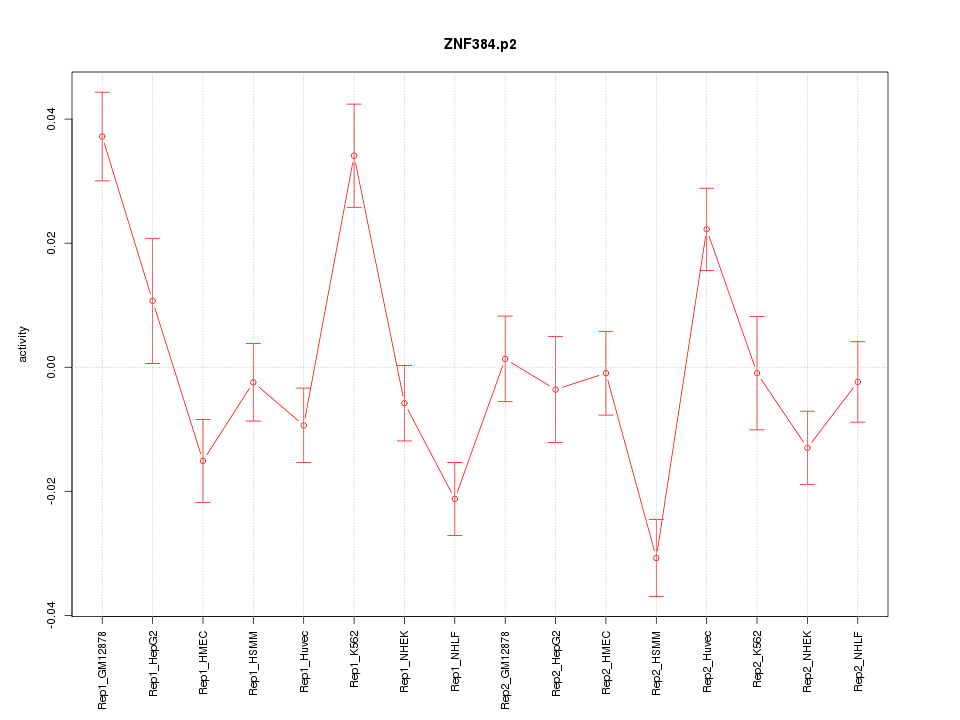
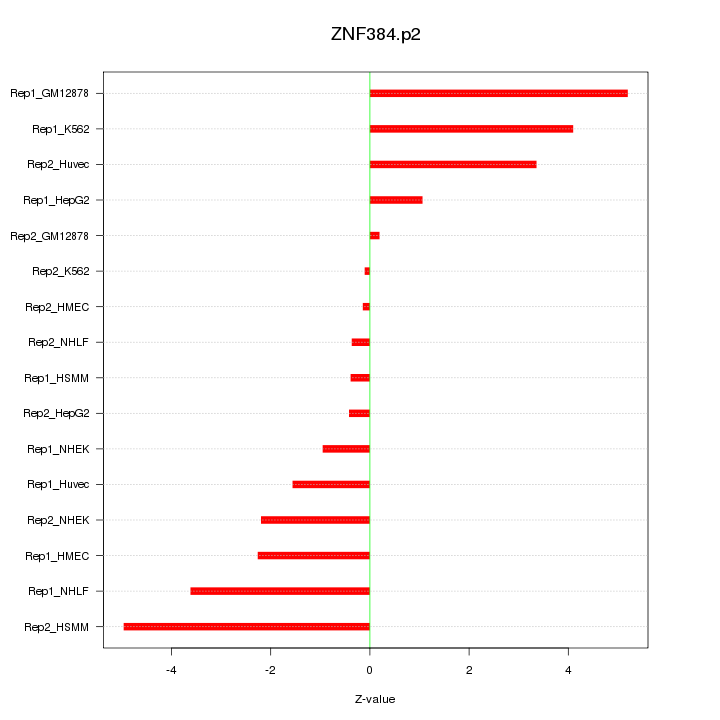
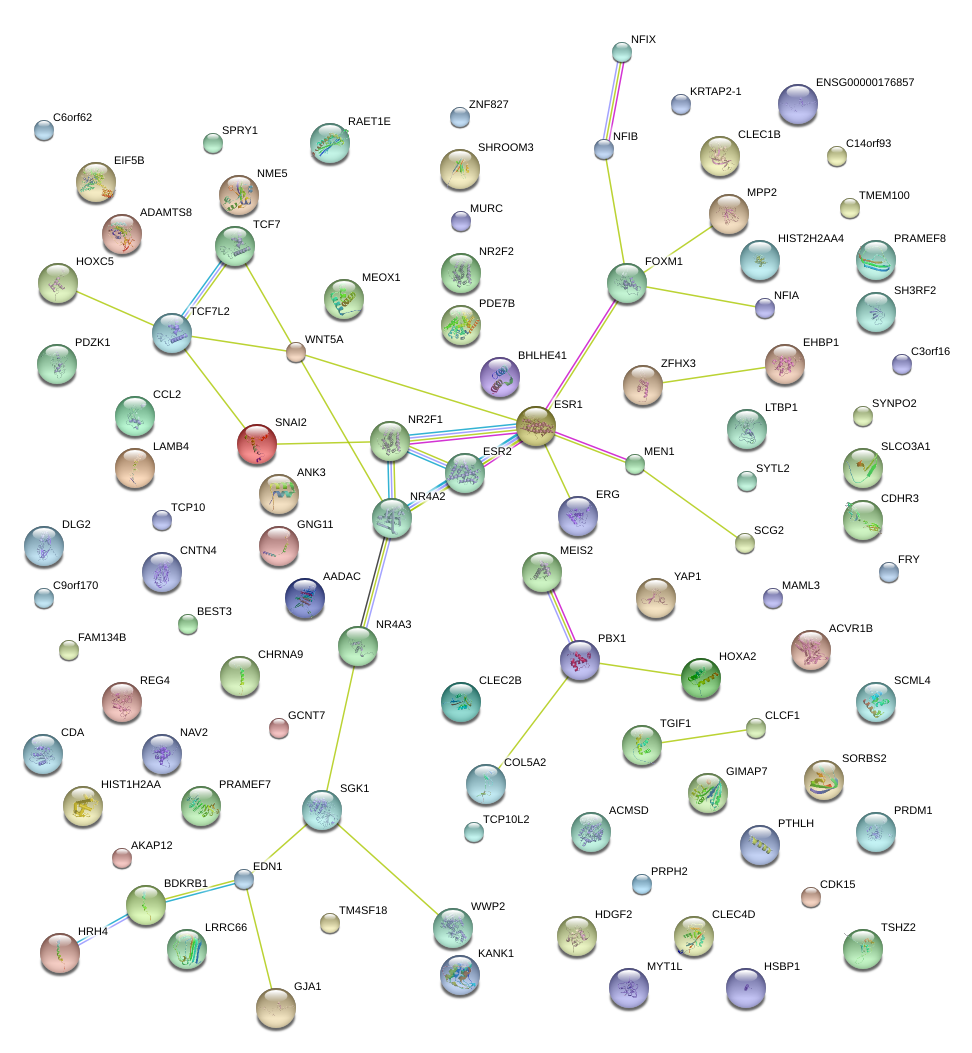
|
chr11_+_101488380
|
2.856
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr6_+_12398514
|
2.797
|
NM_001168319
NM_001955
|
EDN1
|
endothelin 1
|
|
chr3_-_150533948
|
2.633
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr6_+_106653429
|
2.584
|
NM_182907
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr5_-_172016174
|
2.531
|
|
|
|
|
chr4_+_77575276
|
2.251
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr1_+_162795328
|
2.190
|
NM_002585
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr10_-_61819493
|
2.024
|
NM_020987
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr15_+_90198567
|
1.961
|
|
SLCO3A1
|
solute carrier organic anion transporter family, member 3A1
|
|
chr5_+_92944680
|
1.874
|
NM_005654
|
NR2F1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr4_-_52578542
|
1.867
|
NM_001024611
|
LRRC66
|
leucine rich repeat containing 66
|
|
chr21_-_38792173
|
1.836
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr2_-_189752575
|
1.824
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr20_-_54534387
|
1.818
|
NM_080615
|
GCNT7
|
glucosaminyl (N-acetyl) transferase family member 7
|
|
chr17_+_29606408
|
1.696
|
NM_002982
|
CCL2
|
chemokine (C-C motif) ligand 2
|
|
chr6_-_24827265
|
1.665
|
NM_030939
|
C6orf62
|
chromosome 6 open reading frame 62
|
|
chr16_+_68516401
|
1.634
|
NM_199424
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2
|
|
chr1_+_162795538
|
1.610
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr3_-_55489973
|
1.581
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr6_+_151688327
|
1.578
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr4_+_120029426
|
1.577
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr6_+_121798443
|
1.570
|
NM_000165
|
GJA1
|
gap junction protein, alpha 1, 43kDa
|
|
chr18_+_3443771
|
1.520
|
NM_173211
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr7_+_149842850
|
1.513
|
NM_153236
|
GIMAP7
|
GTPase, IMAP family member 7
|
|
chr4_-_147079018
|
1.501
|
NM_178835
|
ZNF827
|
zinc finger protein 827
|
|
chr6_-_134680888
|
1.498
|
NM_001143676
|
SGK1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_26169074
|
1.497
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr7_-_107558021
|
1.482
|
NM_007356
|
LAMB4
|
laminin, beta 4
|
|
chr3_+_3056307
|
1.468
|
NM_175612
|
CNTN4
|
contactin 4
|
|
chr17_-_51155073
|
1.451
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr9_+_88953378
|
1.431
|
NM_001001709
|
C9orf170
|
chromosome 9 open reading frame 170
|
|
chr7_-_27108826
|
1.428
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr10_+_114699953
|
1.420
|
NM_001146274
NM_001146283
NM_001146284
NM_001146285
NM_001146286
NM_001198525
NM_001198526
NM_001198527
NM_001198528
NM_001198529
NM_001198530
NM_001198531
NM_030756
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr3_-_150993299
|
1.419
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr6_+_136214495
|
1.415
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr11_-_85115105
|
1.397
|
NM_206927
NM_206928
|
SYTL2
|
synaptotagmin-like 2
|
|
chr11_+_20000938
|
1.392
|
|
NAV2
|
neuron navigator 2
|
|
chr5_-_137502975
|
1.355
|
NM_003551
|
NME5
|
non-metastatic cells 5, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr6_+_152053323
|
1.342
|
NM_001122742
|
ESR1
|
estrogen receptor 1
|
|
chr2_-_156897286
|
1.340
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr1_+_20788027
|
1.320
|
NM_001785
|
CDA
|
cytidine deaminase
|
|
chr4_-_147079410
|
1.319
|
|
|
|
|
chr4_+_159311353
|
1.295
|
|
LOC285505
|
hypothetical protein LOC285505
|
|
chr6_-_108252200
|
1.286
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr14_-_22549168
|
1.282
|
NM_001130706
NM_001130708
NM_021944
|
C14orf93
|
chromosome 14 open reading frame 93
|
|
chr12_+_52713098
|
1.280
|
NM_018953
|
HOXC5
|
homeobox C5
|
|
chr3_+_153014550
|
1.265
|
NM_001086
|
AADAC
|
arylacetamide deacetylase (esterase)
|
|
chr14_+_95792299
|
1.264
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chr2_+_33213111
|
1.263
|
NM_000627
NM_001166264
NM_001166265
NM_001166266
|
LTBP1
|
latent transforming growth factor beta binding protein 1
|
|
chr2_+_99344060
|
1.191
|
|
EIF5B
|
eukaryotic translation initiation factor 5B
|
|
chr11_-_85015961
|
1.186
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr2_-_224175386
|
1.176
|
NM_003469
|
SCG2
|
secretogranin II
|
|
chr13_+_31503436
|
1.166
|
NM_023037
|
FRY
|
furry homolog (Drosophila)
|
|
chr12_-_28016182
|
1.160
|
NM_002820
NM_198965
|
PTHLH
|
parathyroid hormone-like hormone
|
|
chr6_-_150253729
|
1.155
|
NM_139165
|
RAET1E
|
retinoic acid early transcript 1E
|
|
chr17_-_36469869
|
1.153
|
NM_001165252
|
LOC730755
|
keratin associated protein 2-4-like
|
|
chr7_+_105304445
|
1.151
|
|
LOC100506768
CDHR3
|
hypothetical LOC100506768
cadherin-related family member 3
|
|
chr1_+_61697209
|
1.146
|
|
NFIA
|
nuclear factor I/A
|
|
chr9_+_102380156
|
1.144
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr1_+_12899036
|
1.144
|
NM_001012276
|
PRAMEF8
PRAMEF7
|
PRAME family member 8
PRAME family member 7
|
|
chr12_-_68379331
|
1.128
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr12_+_50633430
|
1.118
|
NM_020327
|
ACVR1B
|
activin A receptor, type IB
|
|
chr12_-_9913199
|
1.112
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr17_-_39094427
|
1.111
|
NM_004527
NM_013999
|
MEOX1
|
mesenchyme homeobox 1
|
|
chr2_+_62754489
|
1.109
|
NM_001142615
|
EHBP1
|
EH domain binding protein 1
|
|
chr15_+_94670160
|
1.109
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr4_+_40032146
|
1.085
|
NM_017581
|
CHRNA9
|
cholinergic receptor, nicotinic, alpha 9
|
|
chr4_-_186934059
|
1.085
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_167504070
|
1.075
|
NM_001145121
|
TCP10L2
TCP10
|
t-complex 10-like 2 (mouse)
t-complex 10 homolog (mouse)
|
|
chr19_+_4442610
|
1.073
|
|
HDGFRP2
|
hepatoma-derived growth factor-related protein 2
|
|
chr5_-_16561936
|
1.059
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chr1_-_120155581
|
1.058
|
NM_001159352
NM_001159353
NM_032044
|
REG4
|
regenerating islet-derived family, member 4
|
|
chr18_+_20294590
|
1.057
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr2_+_202379442
|
1.051
|
NM_139158
|
CDK15
|
cyclin-dependent kinase 15
|
|
chr15_-_35178888
|
1.051
|
NM_172316
|
MEIS2
|
Meis homeobox 2
|
|
chr2_-_224175339
|
1.049
|
|
SCG2
|
secretogranin II
|
|
chr2_+_135312655
|
1.046
|
NM_138326
|
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase
|
|
chr5_-_42847716
|
1.042
|
NM_001085486
NM_001093726
NM_005410
|
SEPP1
|
selenoprotein P, plasma, 1
|
|
chr8_-_49996353
|
1.035
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr20_+_51023159
|
1.026
|
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr12_-_9913724
|
1.022
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr16_-_71650034
|
1.018
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr9_+_696744
|
1.016
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_+_144439069
|
1.008
|
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr4_+_124537334
|
1.008
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr10_-_61570665
|
1.007
|
NM_001149
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G)
|
|
chr11_-_66898223
|
1.007
|
NM_001166212
|
CLCF1
|
cardiotrophin-like cytokine factor 1
|
|
chr7_+_93388951
|
1.001
|
NM_004126
|
GNG11
|
guanine nucleotide binding protein (G protein), gamma 11
|
|
chr12_+_8557402
|
0.996
|
NM_080387
|
CLEC4D
|
C-type lectin domain family 4, member D
|
|
chr6_-_25834733
|
0.990
|
NM_170745
|
HIST1H2AA
|
histone cluster 1, H2aa
|
|
chr4_-_141293572
|
0.984
|
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr1_+_171871283
|
0.984
|
NM_001195190
|
LOC730159
|
hypothetical protein LOC730159
|
|
chr2_-_1905508
|
0.981
|
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr5_+_145296313
|
0.975
|
NM_152550
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr12_+_52733927
|
0.975
|
NM_153633
|
HOXC4
HOXC6
|
homeobox C4
homeobox C6
|
|
chr2_-_224175293
|
0.973
|
|
SCG2
|
secretogranin II
|
|
chr5_-_124108672
|
0.968
|
NM_020747
|
ZNF608
|
zinc finger protein 608
|
|
chr1_-_53972464
|
0.968
|
NM_147193
|
GLIS1
|
GLIS family zinc finger 1
|
|
chr9_-_106705780
|
0.947
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr4_+_154397975
|
0.946
|
|
TRIM2
|
tripartite motif containing 2
|
|
chr1_-_12831164
|
0.942
|
NM_001013631
|
HNRNPCL1
|
heterogeneous nuclear ribonucleoprotein C-like 1
|
|
chr8_+_124032830
|
0.939
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr5_+_140844810
|
0.939
|
NM_018928
NM_032406
|
PCDHGC4
|
protocadherin gamma subfamily C, 4
|
|
chr2_-_37397669
|
0.928
|
NM_005813
|
PRKD3
|
protein kinase D3
|
|
chr11_-_102331643
|
0.925
|
NM_002427
|
MMP13
|
matrix metallopeptidase 13 (collagenase 3)
|
|
chr4_-_87989291
|
0.919
|
NM_197965
|
SLC10A6
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 6
|
|
chr19_+_4720144
|
0.919
|
|
C19orf30
|
chromosome 19 open reading frame 30
|
|
chr1_-_23393808
|
0.917
|
NM_000864
|
HTR1D
|
5-hydroxytryptamine (serotonin) receptor 1D
|
|
chr10_+_114700775
|
0.915
|
|
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box)
|
|
chr1_+_173303616
|
0.914
|
NM_022093
|
TNN
|
tenascin N
|
|
chr1_+_66772393
|
0.912
|
NM_032291
|
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1
|
|
chr6_+_1556543
|
0.908
|
|
FOXC1
|
forkhead box C1
|
|
chr18_-_65775139
|
0.902
|
NM_006566
|
CD226
|
CD226 molecule
|
|
chr9_+_117955890
|
0.899
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr15_-_35179614
|
0.898
|
|
MEIS2
|
Meis homeobox 2
|
|
chr6_-_116973465
|
0.895
|
NM_001139444
|
BET3L
|
BET3 like (S. cerevisiae)
|
|
chr6_+_167624792
|
0.890
|
NM_001143947
NM_018974
|
UNC93A
|
unc-93 homolog A (C. elegans)
|
|
chr7_+_153633127
|
0.885
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr4_+_187227302
|
0.879
|
NM_003265
|
TLR3
|
toll-like receptor 3
|
|
chr7_+_116182676
|
0.874
|
|
|
|
|
chr2_+_101680594
|
0.868
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr12_+_88268845
|
0.868
|
|
LOC100131490
|
hypothetical LOC100131490
|
|
chr14_+_99555471
|
0.862
|
|
EVL
|
Enah/Vasp-like
|
|
chr9_-_106705867
|
0.861
|
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr21_-_34820916
|
0.860
|
NM_203418
|
RCAN1
|
regulator of calcineurin 1
|
|
chr1_+_22107137
|
0.859
|
|
RPL21
|
ribosomal protein L21
|
|
chr5_+_36644187
|
0.858
|
NM_001166695
|
SLC1A3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr5_-_74362479
|
0.856
|
NM_016591
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2
|
|
chr9_-_111230926
|
0.846
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr4_-_141294682
|
0.843
|
NM_018717
|
MAML3
|
mastermind-like 3 (Drosophila)
|
|
chr6_+_106640903
|
0.843
|
|
PRDM1
|
PR domain containing 1, with ZNF domain
|
|
chr20_+_19818209
|
0.843
|
NM_018993
|
RIN2
|
Ras and Rab interactor 2
|
|
chr1_+_242281199
|
0.841
|
|
ZNF238
|
zinc finger protein 238
|
|
chr6_-_139654807
|
0.833
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr14_+_62740878
|
0.832
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr1_-_151280201
|
0.827
|
NM_006945
|
SPRR2D
|
small proline-rich protein 2D
|
|
chr2_-_213723298
|
0.823
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr3_+_113200696
|
0.808
|
NM_001008273
|
TAGLN3
|
transgelin 3
|
|
chr20_-_49747397
|
0.807
|
|
ATP9A
|
ATPase, class II, type 9A
|
|
chr12_-_54392328
|
0.806
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr6_+_74462654
|
0.802
|
|
CD109
|
CD109 molecule
|
|
chr7_+_129720209
|
0.802
|
NM_001163446
NM_016352
|
CPA4
|
carboxypeptidase A4
|
|
chr19_-_56825400
|
0.802
|
NM_003830
|
SIGLEC5
|
sialic acid binding Ig-like lectin 5
|
|
chr11_-_102001259
|
0.802
|
NM_004771
|
MMP20
|
matrix metallopeptidase 20
|
|
chr6_-_46401174
|
0.801
|
|
RCAN2
|
regulator of calcineurin 2
|
|
chr16_+_24475129
|
0.801
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr3_-_191322919
|
0.798
|
NM_001134418
|
LEPREL1
|
leprecan-like 1
|
|
chr15_+_60838053
|
0.797
|
|
TLN2
|
talin 2
|
|
chr12_+_74160779
|
0.795
|
NM_006851
|
GLIPR1
|
GLI pathogenesis-related 1
|
|
chr1_+_163084934
|
0.795
|
|
PBX1
|
pre-B-cell leukemia homeobox 1
|
|
chr1_+_169747788
|
0.792
|
|
PRRC2C
|
proline-rich coiled-coil 2C
|
|
chr4_+_111616630
|
0.780
|
NM_001977
|
ENPEP
|
glutamyl aminopeptidase (aminopeptidase A)
|
|
chr2_-_74424041
|
0.777
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr12_+_27740694
|
0.777
|
NM_001029874
|
REP15
|
RAB15 effector protein
|
|
chr1_-_173979292
|
0.775
|
NM_003285
|
TNR
|
tenascin R (restrictin, janusin)
|
|
chr2_-_162808123
|
0.772
|
NM_004460
|
FAP
|
fibroblast activation protein, alpha
|
|
chr11_-_76676110
|
0.772
|
NM_182833
|
GDPD4
|
glycerophosphodiester phosphodiesterase domain containing 4
|
|
chr5_-_121842680
|
0.767
|
|
|
|
|
chr3_+_150065732
|
0.764
|
NM_001870
|
CPA3
|
carboxypeptidase A3 (mast cell)
|
|
chr7_+_117651947
|
0.762
|
NM_019644
|
ANKRD7
|
ankyrin repeat domain 7
|
|
chr1_-_167663278
|
0.762
|
NM_021179
|
C1orf114
|
chromosome 1 open reading frame 114
|
|
chr12_-_7547552
|
0.757
|
NM_004244
NM_203416
|
CD163
|
CD163 molecule
|
|
chr4_-_159313167
|
0.757
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr18_-_63333487
|
0.750
|
|
DSEL
|
dermatan sulfate epimerase-like
|
|
chr12_+_118256899
|
0.750
|
NM_178499
|
CCDC60
|
coiled-coil domain containing 60
|
|
chr17_-_61253122
|
0.747
|
NM_001037325
|
CCDC46
|
coiled-coil domain containing 46
|
|
chr5_-_37871349
|
0.747
|
NM_199231
|
GDNF
|
glial cell derived neurotrophic factor
|
|
chr5_+_110435288
|
0.742
|
NM_033035
|
TSLP
|
thymic stromal lymphopoietin
|
|
chr6_+_26381122
|
0.738
|
NM_003525
|
HIST1H2BI
|
histone cluster 1, H2bi
|
|
chr17_-_36507900
|
0.738
|
NM_031960
|
KRTAP4-8
|
keratin associated protein 4-8
|
|
chr17_-_21395483
|
0.736
|
NM_001113434
|
C17orf51
|
chromosome 17 open reading frame 51
|
|
chr1_-_155936904
|
0.734
|
NM_052939
|
FCRL3
|
Fc receptor-like 3
|
|
chr13_-_46368968
|
0.732
|
NM_000621
NM_001165947
|
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A
|
|
chr1_+_116455898
|
0.730
|
NM_152367
|
MAB21L3
|
mab-21-like 3 (C. elegans)
|
|
chr6_+_123151669
|
0.726
|
NM_006714
|
SMPDL3A
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr6_+_31016755
|
0.720
|
NM_080870
|
DPCR1
|
diffuse panbronchiolitis critical region 1
|
|
chr15_+_88345612
|
0.720
|
NM_198526
|
ZNF710
|
zinc finger protein 710
|
|
chr2_+_157822355
|
0.719
|
NM_014568
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5)
|
|
chr4_-_177353707
|
0.716
|
NM_144644
|
SPATA4
|
spermatogenesis associated 4
|
|
chr19_+_6981593
|
0.710
|
NM_001136507
|
MBD3L5
|
methyl-CpG-binding domain protein 3-like 5-like
|
|
chr11_+_20001130
|
0.708
|
NM_001111019
|
NAV2
|
neuron navigator 2
|
|
chr17_-_36807322
|
0.706
|
NM_002277
|
KRT31
|
keratin 31
|
|
chr3_-_55490401
|
0.704
|
|
WNT5A
|
wingless-type MMTV integration site family, member 5A
|
|
chr7_+_20008870
|
0.702
|
|
RPL21
|
ribosomal protein L21
|
|
chr3_-_61703850
|
0.700
|
|
|
|
|
chr11_+_6854431
|
0.694
|
NM_207186
|
OR10A4
|
olfactory receptor, family 10, subfamily A, member 4
|
|
chr4_-_100284395
|
0.693
|
NM_000670
|
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide
|
|
chr11_-_85103438
|
0.692
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr18_-_18251808
|
0.689
|
NM_172241
|
CTAGE1
|
cutaneous T-cell lymphoma-associated antigen 1
|
|
chr12_-_51065629
|
0.687
|
NM_033045
|
KRT84
|
keratin 84
|
|
chr7_-_135063461
|
0.682
|
NM_012450
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporters), member 4
|
|
chr7_+_22733626
|
0.682
|
|
IL6
|
interleukin 6 (interferon, beta 2)
|
|
chr17_-_36418869
|
0.681
|
NM_031958
|
KRTAP3-1
|
keratin associated protein 3-1
|
|
chr1_-_116184846
|
0.677
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr6_-_152681171
|
0.674
|
NM_015293
|
SYNE1
|
spectrin repeat containing, nuclear envelope 1
|
|
chr9_-_14076901
|
0.671
|
|
NFIB
|
nuclear factor I/B
|
|
chr15_-_35179185
|
0.671
|
|
MEIS2
|
Meis homeobox 2
|
|
chr18_-_51406841
|
0.669
|
NM_001083962
NM_003199
|
TCF4
|
transcription factor 4
|
|
chr6_+_167456230
|
0.667
|
NM_031409
|
CCR6
|
chemokine (C-C motif) receptor 6
|
|
chr1_+_113734997
|
0.665
|
NM_001142782
NM_152900
|
MAGI3
|
membrane associated guanylate kinase, WW and PDZ domain containing 3
|