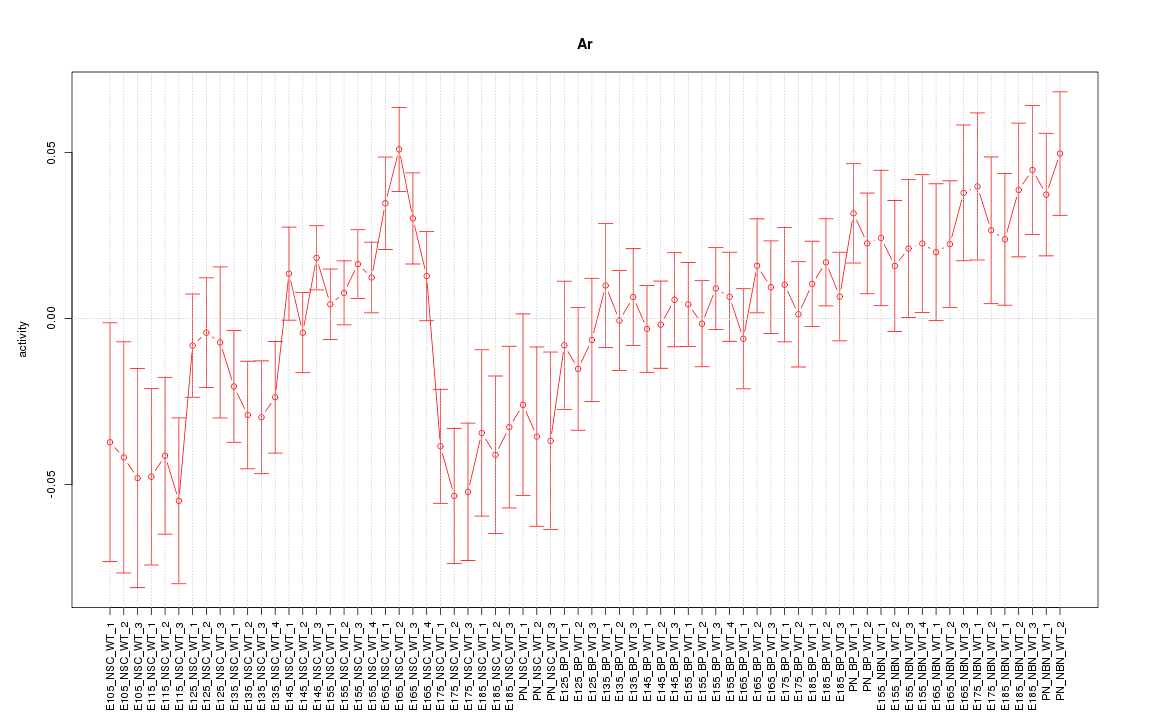
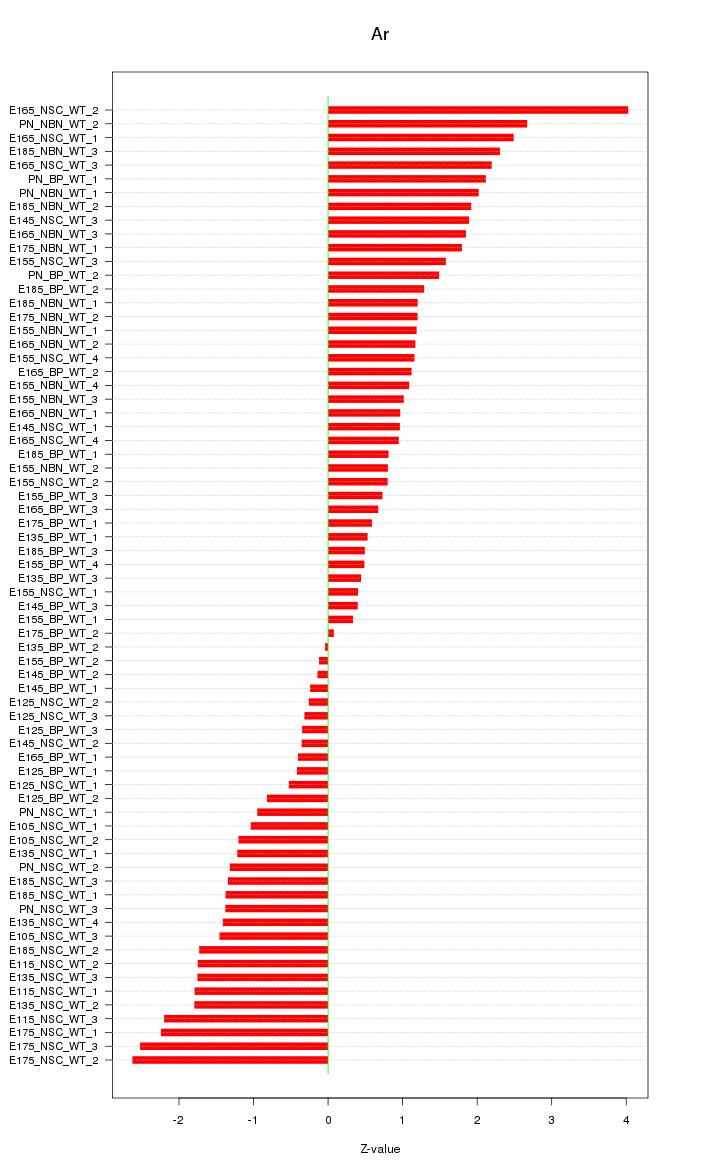
Motif ID: Ar
Z-value: 1.431
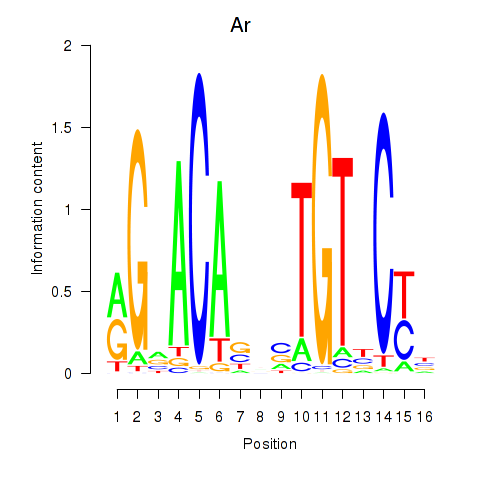
Transcription factors associated with Ar:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ar | ENSMUSG00000046532.7 | Ar |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 39.4 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 2.8 | 8.3 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 2.6 | 7.9 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 2.4 | 11.9 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 2.2 | 9.0 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 2.2 | 10.8 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 1.9 | 5.7 | GO:0010751 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 1.8 | 5.5 | GO:0014877 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 1.8 | 7.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 1.7 | 5.2 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 1.6 | 4.8 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.5 | 4.6 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 1.5 | 10.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 1.2 | 6.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 1.1 | 6.9 | GO:1901524 | regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 1.1 | 12.4 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.0 | 4.2 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 1.0 | 3.0 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.0 | 2.9 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.9 | 6.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.8 | 4.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.8 | 4.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 3.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.8 | 3.8 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.7 | 2.9 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.7 | 4.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.7 | 5.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.7 | 4.0 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 | 1.9 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.6 | 3.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.6 | 29.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) |
| 0.6 | 2.2 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.6 | 6.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.5 | 1.6 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.5 | 5.8 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.5 | 3.5 | GO:0061092 | involuntary skeletal muscle contraction(GO:0003011) regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.5 | 2.0 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.5 | 3.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.5 | 2.9 | GO:0000429 | carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) |
| 0.5 | 1.4 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.5 | 1.4 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.4 | 1.8 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.4 | 1.7 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.4 | 2.8 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.4 | 1.2 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.4 | 5.0 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.3 | 6.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 0.7 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.3 | 1.0 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.3 | 1.0 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.3 | 5.6 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.3 | 10.8 | GO:0009268 | response to pH(GO:0009268) |
| 0.3 | 1.6 | GO:0042167 | heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.3 | 1.9 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.3 | 6.4 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 | 2.5 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 8.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.3 | 8.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.3 | 9.4 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.3 | 2.0 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.3 | 2.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.2 | 7.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.2 | 0.5 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.2 | 4.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.2 | 7.0 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 1.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.2 | 13.1 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.2 | 2.7 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
| 0.2 | 2.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.2 | 3.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 4.1 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.2 | 3.1 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.2 | 3.7 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.2 | 1.2 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 2.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 1.7 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 0.7 | GO:0050904 | diapedesis(GO:0050904) |
| 0.2 | 5.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.2 | 1.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.2 | 0.9 | GO:0071361 | detection of calcium ion(GO:0005513) cellular response to ethanol(GO:0071361) |
| 0.1 | 17.3 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 1.6 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.1 | 0.4 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 4.0 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 1.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.5 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.4 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) negative regulation of protein phosphatase type 2B activity(GO:0032513) |
| 0.1 | 0.7 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.1 | 2.0 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 | 3.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.1 | 0.5 | GO:0090148 | membrane fission(GO:0090148) |
| 0.1 | 8.3 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 0.9 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.4 | GO:0071499 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.1 | 4.7 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 1.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.1 | 1.5 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.4 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 | 1.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.1 | 3.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 1.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.1 | 3.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.1 | 1.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.1 | 3.1 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.2 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.1 | 2.3 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 2.8 | GO:0001707 | mesoderm formation(GO:0001707) |
| 0.1 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.1 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 0.4 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 5.7 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.8 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.4 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 1.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 4.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.5 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.0 | 1.6 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 | 1.0 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 2.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 3.2 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.4 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 0.3 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 1.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.6 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 43.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 1.2 | 6.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 1.2 | 10.8 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.1 | 5.6 | GO:0000798 | nuclear cohesin complex(GO:0000798) nuclear meiotic cohesin complex(GO:0034991) |
| 1.0 | 28.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.9 | 3.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.8 | 4.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.7 | 2.7 | GO:0002141 | stereocilia ankle link(GO:0002141) |
| 0.7 | 8.0 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.6 | 2.6 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.5 | 3.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.4 | 20.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.3 | 2.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.5 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.3 | 5.7 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.3 | 1.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.3 | 4.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 2.6 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 4.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 2.4 | GO:0030867 | desmosome(GO:0030057) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 4.8 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 2.1 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.2 | 2.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.2 | 6.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.6 | GO:0042581 | specific granule(GO:0042581) |
| 0.1 | 2.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 0.7 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.8 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 3.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.1 | 7.7 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 5.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 4.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 3.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 7.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 8.0 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 1.1 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 2.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 0.2 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.1 | 2.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.1 | 5.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.2 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 3.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.5 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 3.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 1.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 5.0 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 5.0 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.6 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.9 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 5.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 19.4 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 14.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 4.1 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.5 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.9 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 3.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 2.7 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.0 | 1.8 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.0 | 29.9 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 5.6 | 28.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 2.3 | 6.9 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 2.2 | 6.7 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 2.1 | 8.3 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 1.7 | 5.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 1.6 | 8.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.5 | 4.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.5 | 4.6 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 1.3 | 6.4 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 1.2 | 7.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 1.1 | 6.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 1.0 | 31.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 1.0 | 10.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.9 | 3.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.8 | 4.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.8 | 3.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.8 | 6.0 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.7 | 4.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.6 | 5.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.6 | 7.0 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.6 | 2.2 | GO:0023029 | peptide antigen-transporting ATPase activity(GO:0015433) MHC class Ib protein binding(GO:0023029) tapasin binding(GO:0046980) |
| 0.5 | 2.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.5 | 4.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.5 | 7.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.5 | 2.9 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.5 | 2.4 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.5 | 2.9 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.5 | 10.8 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 5.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.4 | 6.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.4 | 1.6 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.4 | 1.6 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 1.2 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 2.3 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.4 | 1.4 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.3 | 1.7 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.3 | 1.7 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.3 | 7.7 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 3.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.3 | 3.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.3 | 1.8 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.3 | 2.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.3 | 7.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 9.6 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 1.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 1.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.2 | 2.0 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.6 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.2 | 7.0 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.2 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 12.0 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.2 | 3.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.2 | 0.7 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.2 | 0.5 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 1.0 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.1 | 2.8 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.9 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 1.9 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 8.5 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 5.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.5 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 5.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 7.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 4.5 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.1 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 1.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 8.3 | GO:0002039 | p53 binding(GO:0002039) |
| 0.1 | 2.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 3.8 | GO:0045502 | dynein binding(GO:0045502) |
| 0.1 | 1.5 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 12.3 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 12.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 2.9 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 13.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.3 | GO:0034511 | U4 snRNA binding(GO:0030621) U3 snoRNA binding(GO:0034511) |
| 0.1 | 2.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 2.8 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 4.3 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.1 | 1.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 3.0 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.4 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 2.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.0 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 2.9 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 2.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.6 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 1.6 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.6 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |