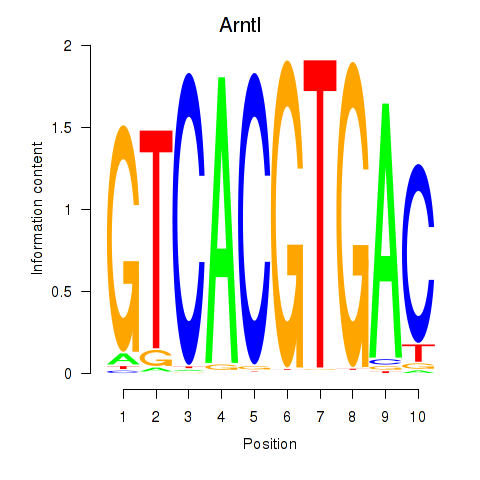
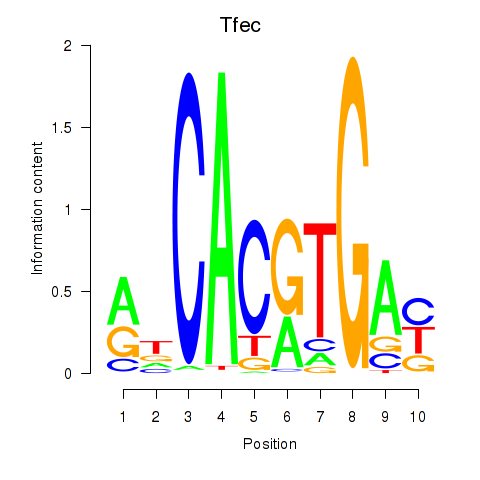
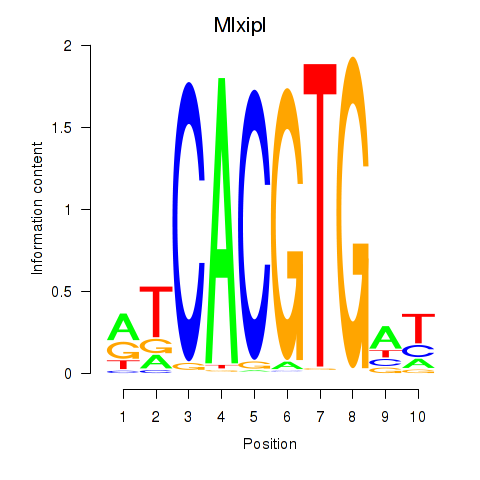
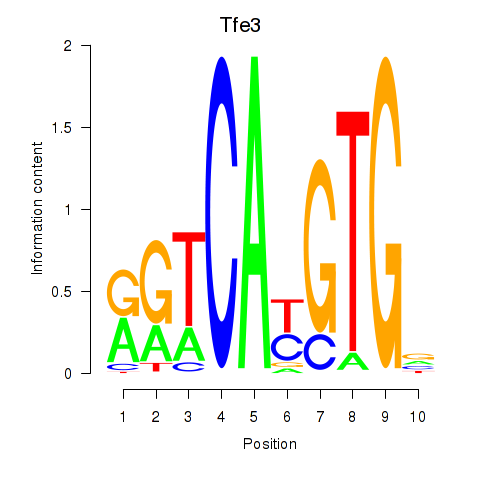
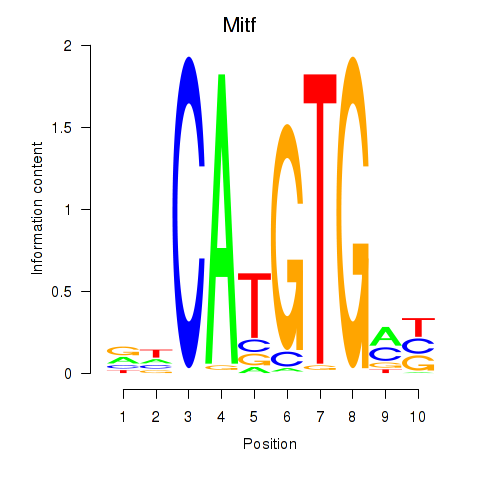
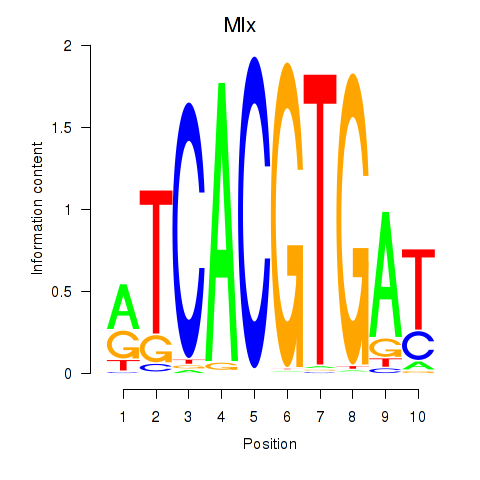
| 1.6 |
6.4 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 1.2 |
4.8 |
GO:0060729 |
intestinal epithelial structure maintenance(GO:0060729) |
| 1.2 |
3.5 |
GO:0010845 |
positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 1.1 |
3.3 |
GO:1902445 |
regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 1.1 |
3.2 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.8 |
2.3 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.8 |
3.1 |
GO:0042364 |
water-soluble vitamin biosynthetic process(GO:0042364) |
| 0.7 |
2.2 |
GO:0032053 |
ciliary basal body organization(GO:0032053) |
| 0.7 |
7.0 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.7 |
2.1 |
GO:0032915 |
positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.7 |
2.1 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.7 |
2.1 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.7 |
5.3 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.7 |
3.3 |
GO:0014022 |
neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.6 |
3.2 |
GO:0035984 |
response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 0.6 |
2.4 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.5 |
3.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.5 |
1.5 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.5 |
1.5 |
GO:0071873 |
response to norepinephrine(GO:0071873) |
| 0.5 |
15.6 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.5 |
2.0 |
GO:0090238 |
positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.5 |
5.3 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 |
1.4 |
GO:0070476 |
rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.5 |
1.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.4 |
1.3 |
GO:0072070 |
establishment or maintenance of polarity of embryonic epithelium(GO:0016332) loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.4 |
1.8 |
GO:0006407 |
rRNA export from nucleus(GO:0006407) |
| 0.4 |
1.3 |
GO:2000508 |
regulation of dendritic cell chemotaxis(GO:2000508) positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.4 |
2.1 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.4 |
1.7 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.4 |
2.1 |
GO:1905049 |
negative regulation of metalloendopeptidase activity(GO:1904684) negative regulation of metallopeptidase activity(GO:1905049) |
| 0.4 |
1.2 |
GO:2000813 |
negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.4 |
1.2 |
GO:0008612 |
peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.4 |
1.2 |
GO:0035864 |
response to potassium ion(GO:0035864) cellular response to potassium ion(GO:0035865) |
| 0.4 |
1.5 |
GO:0010891 |
negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.4 |
1.1 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.4 |
1.8 |
GO:0045627 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.3 |
1.0 |
GO:0060785 |
regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.3 |
3.6 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.3 |
0.6 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.3 |
6.3 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.3 |
0.9 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.3 |
1.2 |
GO:0031622 |
positive regulation of fever generation(GO:0031622) |
| 0.3 |
0.9 |
GO:1902310 |
positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.3 |
3.3 |
GO:0070131 |
positive regulation of mitochondrial translation(GO:0070131) |
| 0.3 |
0.9 |
GO:0060672 |
epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 |
0.8 |
GO:0001732 |
formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.3 |
2.2 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.3 |
0.8 |
GO:0036091 |
positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.3 |
0.3 |
GO:0042726 |
flavin-containing compound metabolic process(GO:0042726) |
| 0.3 |
0.8 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.3 |
0.8 |
GO:1903774 |
positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.3 |
1.6 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.3 |
1.3 |
GO:0051151 |
negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.3 |
0.3 |
GO:0051902 |
negative regulation of mitochondrial depolarization(GO:0051902) negative regulation of membrane depolarization(GO:1904180) |
| 0.3 |
1.3 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.2 |
2.5 |
GO:0061470 |
T follicular helper cell differentiation(GO:0061470) |
| 0.2 |
3.7 |
GO:0002115 |
store-operated calcium entry(GO:0002115) |
| 0.2 |
1.2 |
GO:0043985 |
histone H4-R3 methylation(GO:0043985) |
| 0.2 |
1.4 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.2 |
0.7 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 0.2 |
4.6 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.2 |
1.6 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
3.6 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.2 |
0.4 |
GO:1904058 |
positive regulation of sensory perception of pain(GO:1904058) |
| 0.2 |
1.5 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.2 |
2.6 |
GO:0010807 |
regulation of synaptic vesicle priming(GO:0010807) |
| 0.2 |
1.1 |
GO:0070129 |
regulation of mitochondrial translation(GO:0070129) |
| 0.2 |
1.9 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 |
0.6 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.2 |
1.0 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 |
1.0 |
GO:0040038 |
polar body extrusion after meiotic divisions(GO:0040038) |
| 0.2 |
0.4 |
GO:1904749 |
regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.2 |
1.2 |
GO:0010917 |
negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.2 |
1.2 |
GO:0046502 |
uroporphyrinogen III metabolic process(GO:0046502) |
| 0.2 |
0.6 |
GO:0007181 |
transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 |
3.1 |
GO:1901741 |
positive regulation of myoblast fusion(GO:1901741) |
| 0.2 |
0.6 |
GO:1904742 |
regulation of telomeric DNA binding(GO:1904742) |
| 0.2 |
0.7 |
GO:2001245 |
regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.2 |
0.5 |
GO:1900377 |
negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.2 |
0.5 |
GO:0006669 |
sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.2 |
1.1 |
GO:0002634 |
regulation of germinal center formation(GO:0002634) |
| 0.2 |
2.1 |
GO:0060019 |
radial glial cell differentiation(GO:0060019) |
| 0.2 |
1.7 |
GO:0015824 |
proline transport(GO:0015824) |
| 0.2 |
0.5 |
GO:0006419 |
alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 |
0.7 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.2 |
1.0 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 0.2 |
0.3 |
GO:0060481 |
lobar bronchus epithelium development(GO:0060481) |
| 0.2 |
2.6 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
0.5 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.2 |
0.8 |
GO:0018002 |
N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.2 |
0.8 |
GO:0003356 |
regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.2 |
1.5 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 |
0.9 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.2 |
0.6 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.2 |
0.9 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.2 |
0.8 |
GO:0045716 |
positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.2 |
0.5 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 |
0.3 |
GO:2000643 |
positive regulation of vacuolar transport(GO:1903337) positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.2 |
0.5 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
0.8 |
GO:2000574 |
regulation of microtubule motor activity(GO:2000574) |
| 0.1 |
1.6 |
GO:0032096 |
negative regulation of response to food(GO:0032096) negative regulation of appetite(GO:0032099) |
| 0.1 |
0.4 |
GO:0001978 |
regulation of systemic arterial blood pressure by carotid sinus baroreceptor feedback(GO:0001978) |
| 0.1 |
4.2 |
GO:0009303 |
rRNA transcription(GO:0009303) |
| 0.1 |
0.6 |
GO:0035752 |
lysosomal lumen pH elevation(GO:0035752) |
| 0.1 |
0.1 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 |
2.4 |
GO:0051974 |
negative regulation of telomerase activity(GO:0051974) |
| 0.1 |
0.1 |
GO:1902172 |
keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.1 |
0.8 |
GO:1903071 |
positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 |
0.8 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.1 |
2.0 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.1 |
1.1 |
GO:0035871 |
protein K11-linked deubiquitination(GO:0035871) |
| 0.1 |
0.4 |
GO:0060160 |
negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.1 |
1.3 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.1 |
2.1 |
GO:0061462 |
protein localization to lysosome(GO:0061462) |
| 0.1 |
0.5 |
GO:2000795 |
lung ciliated cell differentiation(GO:0061141) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 |
0.6 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.1 |
1.1 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 |
1.3 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.7 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 |
0.7 |
GO:0002904 |
positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 |
0.4 |
GO:0051866 |
general adaptation syndrome(GO:0051866) |
| 0.1 |
0.6 |
GO:0060620 |
regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.1 |
1.0 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.1 |
0.1 |
GO:0045976 |
negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.1 |
0.8 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.1 |
2.2 |
GO:0097576 |
vacuole fusion(GO:0097576) |
| 0.1 |
0.1 |
GO:0072425 |
signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 |
0.7 |
GO:1904417 |
regulation of xenophagy(GO:1904415) positive regulation of xenophagy(GO:1904417) |
| 0.1 |
0.7 |
GO:0042590 |
antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 |
0.2 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.1 |
0.3 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.1 |
0.3 |
GO:0034287 |
detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 |
0.3 |
GO:0060084 |
synaptic transmission involved in micturition(GO:0060084) |
| 0.1 |
1.2 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
1.0 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.1 |
0.4 |
GO:0006867 |
asparagine transport(GO:0006867) glutamine transport(GO:0006868) |
| 0.1 |
0.3 |
GO:0036353 |
histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 |
0.7 |
GO:0051534 |
negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 |
2.1 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.1 |
0.4 |
GO:0002949 |
tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 |
0.6 |
GO:1903441 |
protein localization to ciliary membrane(GO:1903441) |
| 0.1 |
0.3 |
GO:0006295 |
nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 |
0.7 |
GO:1902035 |
positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.1 |
2.7 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.1 |
1.7 |
GO:0097284 |
hepatocyte apoptotic process(GO:0097284) |
| 0.1 |
0.4 |
GO:0015786 |
UDP-glucose transport(GO:0015786) |
| 0.1 |
0.6 |
GO:0000414 |
regulation of histone H3-K36 methylation(GO:0000414) |
| 0.1 |
0.9 |
GO:0090232 |
positive regulation of spindle checkpoint(GO:0090232) positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.1 |
1.1 |
GO:0030011 |
maintenance of cell polarity(GO:0030011) |
| 0.1 |
0.4 |
GO:0006564 |
L-serine biosynthetic process(GO:0006564) |
| 0.1 |
1.2 |
GO:1990403 |
embryonic brain development(GO:1990403) |
| 0.1 |
0.7 |
GO:2000480 |
regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 |
0.9 |
GO:0032876 |
negative regulation of DNA endoreduplication(GO:0032876) |
| 0.1 |
0.9 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
6.3 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
0.5 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 |
0.3 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
0.6 |
GO:0003222 |
ventricular trabecula myocardium morphogenesis(GO:0003222) |
| 0.1 |
0.2 |
GO:0001777 |
T cell homeostatic proliferation(GO:0001777) |
| 0.1 |
0.4 |
GO:0032959 |
inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 |
0.6 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.1 |
0.5 |
GO:0006123 |
mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 |
0.3 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 0.1 |
0.3 |
GO:0035609 |
C-terminal protein deglutamylation(GO:0035609) |
| 0.1 |
0.2 |
GO:0071677 |
positive regulation of mononuclear cell migration(GO:0071677) |
| 0.1 |
0.4 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
0.3 |
GO:0045607 |
regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 |
0.2 |
GO:0043379 |
memory T cell differentiation(GO:0043379) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 |
0.2 |
GO:0006990 |
positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 |
0.2 |
GO:0071930 |
negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 |
0.2 |
GO:0090220 |
meiotic telomere clustering(GO:0045141) chromosome localization to nuclear envelope involved in homologous chromosome segregation(GO:0090220) |
| 0.1 |
0.5 |
GO:0008616 |
queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.1 |
0.9 |
GO:0006817 |
phosphate ion transport(GO:0006817) |
| 0.1 |
1.6 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
0.2 |
GO:0030421 |
defecation(GO:0030421) |
| 0.1 |
1.3 |
GO:0071786 |
endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 |
0.4 |
GO:0009098 |
branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.1 |
0.4 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 |
0.4 |
GO:0060665 |
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 |
1.0 |
GO:0060837 |
blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 |
0.9 |
GO:0050901 |
leukocyte tethering or rolling(GO:0050901) |
| 0.1 |
0.4 |
GO:0090073 |
positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 |
0.1 |
GO:0045583 |
regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 |
0.1 |
GO:0050748 |
negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.1 |
0.6 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.1 |
0.3 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.6 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
0.5 |
GO:0006089 |
lactate metabolic process(GO:0006089) |
| 0.1 |
0.1 |
GO:0002925 |
positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 |
1.4 |
GO:0002052 |
positive regulation of neuroblast proliferation(GO:0002052) |
| 0.1 |
1.8 |
GO:0048009 |
insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.1 |
0.3 |
GO:0036324 |
vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 |
0.9 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.1 |
1.1 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.1 |
0.6 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.2 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 |
0.4 |
GO:0070474 |
positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 |
0.3 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.1 |
0.4 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 |
0.1 |
GO:0090148 |
membrane fission(GO:0090148) |
| 0.1 |
1.4 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 |
0.3 |
GO:0032298 |
positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 |
0.8 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.2 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.5 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.1 |
0.2 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.1 |
0.1 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
3.9 |
GO:0048477 |
oogenesis(GO:0048477) |
| 0.1 |
0.4 |
GO:0071442 |
positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 |
1.1 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.1 |
0.5 |
GO:0044387 |
negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.1 |
0.2 |
GO:1903800 |
positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 |
0.2 |
GO:0072610 |
interleukin-12 secretion(GO:0072610) regulation of interleukin-12 secretion(GO:2001182) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.1 |
0.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.1 |
0.5 |
GO:0001516 |
prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.1 |
0.8 |
GO:2000279 |
negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.1 |
0.2 |
GO:0070317 |
negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 |
0.2 |
GO:0002337 |
B-1a B cell differentiation(GO:0002337) |
| 0.1 |
0.7 |
GO:1990126 |
retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.1 |
0.2 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.1 |
0.7 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.1 |
0.2 |
GO:0097151 |
positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.1 |
1.0 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.1 |
0.5 |
GO:0010764 |
negative regulation of fibroblast migration(GO:0010764) |
| 0.1 |
0.1 |
GO:0048496 |
maintenance of organ identity(GO:0048496) |
| 0.1 |
0.1 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.1 |
1.0 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
0.5 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.1 |
1.6 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.1 |
0.1 |
GO:0002309 |
T cell proliferation involved in immune response(GO:0002309) |
| 0.1 |
0.6 |
GO:0006283 |
transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 |
0.2 |
GO:0015870 |
acetylcholine transport(GO:0015870) acetate ester transport(GO:1901374) |
| 0.1 |
0.2 |
GO:0070944 |
neutrophil mediated killing of bacterium(GO:0070944) |
| 0.1 |
0.2 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.1 |
0.4 |
GO:0010880 |
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.1 |
0.2 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 0.1 |
0.3 |
GO:0007603 |
phototransduction, visible light(GO:0007603) |
| 0.1 |
1.3 |
GO:0010738 |
regulation of protein kinase A signaling(GO:0010738) |
| 0.1 |
0.1 |
GO:0060743 |
estrous cycle(GO:0044849) epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 |
0.0 |
GO:1901033 |
positive regulation of response to reactive oxygen species(GO:1901033) positive regulation of hydrogen peroxide-mediated programmed cell death(GO:1901300) positive regulation of hydrogen peroxide-induced cell death(GO:1905206) |
| 0.0 |
0.1 |
GO:1900747 |
negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 |
0.3 |
GO:0060346 |
bone trabecula formation(GO:0060346) |
| 0.0 |
0.1 |
GO:0043096 |
purine nucleobase salvage(GO:0043096) |
| 0.0 |
0.9 |
GO:0046856 |
phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 |
0.2 |
GO:0050651 |
dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 |
0.2 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.0 |
0.3 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.0 |
1.7 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.0 |
0.0 |
GO:0098501 |
polynucleotide dephosphorylation(GO:0098501) |
| 0.0 |
0.1 |
GO:0006041 |
glucosamine metabolic process(GO:0006041) |
| 0.0 |
0.1 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 |
0.7 |
GO:0035414 |
negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 |
0.2 |
GO:0061684 |
chaperone-mediated autophagy(GO:0061684) |
| 0.0 |
0.1 |
GO:1901254 |
positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 |
1.0 |
GO:0007520 |
myoblast fusion(GO:0007520) |
| 0.0 |
0.2 |
GO:0051531 |
NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 |
0.2 |
GO:0060373 |
regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 |
0.8 |
GO:0060669 |
embryonic placenta morphogenesis(GO:0060669) |
| 0.0 |
1.5 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.0 |
1.5 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.2 |
GO:0051684 |
maintenance of Golgi location(GO:0051684) |
| 0.0 |
0.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 |
0.1 |
GO:1901859 |
negative regulation of mitochondrial DNA metabolic process(GO:1901859) |
| 0.0 |
0.3 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.0 |
0.3 |
GO:0031000 |
response to caffeine(GO:0031000) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.1 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.0 |
0.3 |
GO:2000767 |
positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 |
1.3 |
GO:0045104 |
intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 |
0.4 |
GO:0031573 |
intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 |
0.5 |
GO:0002385 |
mucosal immune response(GO:0002385) |
| 0.0 |
0.2 |
GO:0070314 |
G1 to G0 transition(GO:0070314) |
| 0.0 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
1.0 |
GO:0018208 |
peptidyl-proline modification(GO:0018208) |
| 0.0 |
0.3 |
GO:0006349 |
regulation of gene expression by genetic imprinting(GO:0006349) DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.8 |
GO:0070584 |
mitochondrion morphogenesis(GO:0070584) |
| 0.0 |
0.0 |
GO:0033092 |
positive regulation of immature T cell proliferation(GO:0033091) positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 |
0.2 |
GO:0000379 |
tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 |
0.3 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.0 |
0.4 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.2 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.0 |
0.9 |
GO:0048246 |
macrophage chemotaxis(GO:0048246) |
| 0.0 |
0.1 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 |
0.2 |
GO:0000480 |
endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 |
0.1 |
GO:1904491 |
protein localization to ciliary transition zone(GO:1904491) |
| 0.0 |
1.9 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.8 |
GO:0048026 |
positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 |
0.2 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.4 |
GO:0032402 |
melanosome transport(GO:0032402) |
| 0.0 |
0.2 |
GO:0046950 |
cellular ketone body metabolic process(GO:0046950) |
| 0.0 |
0.1 |
GO:0060368 |
Fc receptor mediated stimulatory signaling pathway(GO:0002431) regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 |
1.0 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.3 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.1 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 |
0.2 |
GO:0006537 |
glutamate biosynthetic process(GO:0006537) |
| 0.0 |
0.1 |
GO:0070309 |
lens fiber cell morphogenesis(GO:0070309) |
| 0.0 |
0.2 |
GO:0045072 |
regulation of interferon-gamma biosynthetic process(GO:0045072) positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 |
0.8 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.1 |
GO:1902969 |
mitotic DNA replication(GO:1902969) |
| 0.0 |
0.1 |
GO:0086023 |
adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 |
0.2 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 |
0.2 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 0.0 |
0.2 |
GO:0001842 |
neural fold formation(GO:0001842) |
| 0.0 |
0.1 |
GO:0033085 |
negative regulation of T cell differentiation in thymus(GO:0033085) |
| 0.0 |
0.1 |
GO:0046294 |
formaldehyde catabolic process(GO:0046294) |
| 0.0 |
2.2 |
GO:0043473 |
pigmentation(GO:0043473) |
| 0.0 |
0.1 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 |
0.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.4 |
GO:1900047 |
negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 |
0.4 |
GO:0006895 |
Golgi to endosome transport(GO:0006895) |
| 0.0 |
0.4 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 |
0.4 |
GO:0050849 |
negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 |
0.1 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.0 |
0.1 |
GO:0090035 |
regulation of chaperone-mediated protein complex assembly(GO:0090034) positive regulation of chaperone-mediated protein complex assembly(GO:0090035) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.7 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
0.1 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.0 |
0.3 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 |
0.9 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.5 |
GO:0035025 |
positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 |
0.2 |
GO:0071850 |
mitotic cell cycle arrest(GO:0071850) |
| 0.0 |
1.0 |
GO:0034724 |
DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 |
0.2 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
2.2 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.1 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.0 |
1.4 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.0 |
0.1 |
GO:1902902 |
negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 |
0.3 |
GO:0002689 |
negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 |
0.1 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 |
0.1 |
GO:0090529 |
barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 |
0.1 |
GO:0000479 |
endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 |
0.1 |
GO:0042891 |
antibiotic transport(GO:0042891) |
| 0.0 |
0.2 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 |
0.2 |
GO:0051446 |
positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 |
0.6 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.0 |
0.3 |
GO:2000463 |
positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 |
0.7 |
GO:0048747 |
muscle fiber development(GO:0048747) |
| 0.0 |
0.1 |
GO:2000291 |
regulation of myoblast proliferation(GO:2000291) |
| 0.0 |
0.1 |
GO:0045213 |
neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 |
0.1 |
GO:1900118 |
negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 |
0.2 |
GO:0006957 |
complement activation, alternative pathway(GO:0006957) |
| 0.0 |
0.0 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 |
0.1 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 |
0.1 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.0 |
0.6 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
0.3 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.0 |
0.5 |
GO:0042149 |
cellular response to glucose starvation(GO:0042149) |
| 0.0 |
0.1 |
GO:0010988 |
regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 |
0.6 |
GO:0071158 |
positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 |
0.1 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.0 |
0.1 |
GO:0042769 |
DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 |
0.6 |
GO:0006289 |
nucleotide-excision repair(GO:0006289) |
| 0.0 |
0.1 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 |
0.1 |
GO:0090234 |
regulation of kinetochore assembly(GO:0090234) |
| 0.0 |
0.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
1.1 |
GO:0010501 |
RNA secondary structure unwinding(GO:0010501) |
| 0.0 |
0.4 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.3 |
GO:1901409 |
regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 |
0.4 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.7 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.0 |
0.1 |
GO:0048208 |
vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.0 |
0.1 |
GO:0075525 |
viral translational termination-reinitiation(GO:0075525) |
| 0.0 |
0.1 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 |
1.6 |
GO:0006888 |
ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 |
0.1 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 |
0.2 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 |
0.2 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.0 |
0.7 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.2 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.0 |
0.2 |
GO:0045292 |
mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 |
0.1 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 |
0.1 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.9 |
GO:1902668 |
negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 |
0.1 |
GO:2000671 |
regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 |
0.4 |
GO:0043248 |
proteasome assembly(GO:0043248) |
| 0.0 |
0.4 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.0 |
0.5 |
GO:0001508 |
action potential(GO:0001508) |
| 0.0 |
0.1 |
GO:0044337 |
canonical Wnt signaling pathway involved in positive regulation of apoptotic process(GO:0044337) |
| 0.0 |
0.2 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.0 |
0.1 |
GO:0045956 |
positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 |
0.1 |
GO:1903551 |
regulation of extracellular exosome assembly(GO:1903551) |
| 0.0 |
0.8 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |
| 0.0 |
0.5 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.0 |
0.3 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.0 |
0.2 |
GO:0034497 |
protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 |
1.2 |
GO:0015992 |
proton transport(GO:0015992) |
| 0.0 |
0.2 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 |
0.1 |
GO:0070966 |
nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 |
0.2 |
GO:0031665 |
negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 |
0.1 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 |
0.2 |
GO:0048011 |
neurotrophin TRK receptor signaling pathway(GO:0048011) |
| 0.0 |
0.3 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.4 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.0 |
0.5 |
GO:0009268 |
response to pH(GO:0009268) |
| 0.0 |
0.5 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.0 |
0.2 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
0.3 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 |
0.1 |
GO:0033623 |
regulation of integrin activation(GO:0033623) |
| 0.0 |
0.4 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.1 |
GO:0006983 |
ER overload response(GO:0006983) |
| 0.0 |
0.1 |
GO:0000429 |
carbon catabolite regulation of transcription from RNA polymerase II promoter(GO:0000429) carbon catabolite activation of transcription from RNA polymerase II promoter(GO:0000436) carbon catabolite activation of transcription(GO:0045991) |
| 0.0 |
0.2 |
GO:0019730 |
antimicrobial humoral response(GO:0019730) |
| 0.0 |
0.3 |
GO:0009048 |
dosage compensation(GO:0007549) dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 |
0.5 |
GO:0042773 |
ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 |
0.1 |
GO:0060716 |
labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 |
0.2 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.0 |
0.1 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 |
0.1 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 |
0.1 |
GO:0031648 |
protein destabilization(GO:0031648) |
| 0.0 |
0.1 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 |
0.6 |
GO:0032088 |
negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 |
0.2 |
GO:0007588 |
excretion(GO:0007588) |
| 0.0 |
0.2 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.0 |
0.5 |
GO:0000187 |
activation of MAPK activity(GO:0000187) |
| 0.0 |
0.1 |
GO:0007168 |
receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 |
0.1 |
GO:0060363 |
cranial suture morphogenesis(GO:0060363) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.3 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.1 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.0 |
0.3 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.1 |
GO:1904263 |
positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 |
0.0 |
GO:0018022 |
peptidyl-lysine methylation(GO:0018022) |
| 0.0 |
0.1 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 |
0.2 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 |
0.3 |
GO:0019233 |
sensory perception of pain(GO:0019233) |
| 0.0 |
0.2 |
GO:0071364 |
cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 |
0.1 |
GO:0050853 |
B cell receptor signaling pathway(GO:0050853) |
| 0.0 |
0.0 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.0 |
0.0 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 |
0.1 |
GO:0002507 |
tolerance induction(GO:0002507) |
| 0.0 |
0.0 |
GO:0043983 |
histone H4-K12 acetylation(GO:0043983) |
| 0.0 |
0.4 |
GO:0006342 |
chromatin silencing(GO:0006342) negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 |
0.1 |
GO:0031119 |
tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 |
0.1 |
GO:0021554 |
optic nerve development(GO:0021554) |
| 0.0 |
0.3 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.2 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
0.1 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 |
0.1 |
GO:0090502 |
RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 |
0.4 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.1 |
GO:0072321 |
chaperone-mediated protein transport(GO:0072321) |
| 0.0 |
0.1 |
GO:0006108 |
malate metabolic process(GO:0006108) |
| 0.0 |
0.1 |
GO:0034776 |
response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.0 |
0.0 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
0.1 |
GO:0061732 |
mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 |
0.1 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.1 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 |
0.1 |
GO:0002024 |
diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 |
0.1 |
GO:0044068 |
modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 |
0.2 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.0 |
1.5 |
GO:0016579 |
protein deubiquitination(GO:0016579) |
| 0.0 |
0.1 |
GO:0070234 |
positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 |
0.1 |
GO:1900746 |
regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.0 |
0.1 |
GO:0061003 |
positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 |
0.2 |
GO:0030513 |
positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 |
1.0 |
GO:1903749 |
positive regulation of establishment of protein localization to mitochondrion(GO:1903749) positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.1 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 |
0.2 |
GO:0043982 |
histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 |
0.3 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
0.3 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.1 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 |
0.0 |
GO:0006582 |
melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 |
0.7 |
GO:0032436 |
positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 |
0.0 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 |
0.0 |
GO:1900126 |
negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 |
0.0 |
GO:1903977 |
positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.0 |
0.1 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.0 |
GO:0003085 |
negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 |
0.1 |
GO:0007190 |
activation of adenylate cyclase activity(GO:0007190) |
| 0.0 |
0.1 |
GO:0006501 |
C-terminal protein lipidation(GO:0006501) |
| 0.0 |
0.1 |
GO:0002335 |
mature B cell differentiation(GO:0002335) |
| 0.0 |
0.1 |
GO:0051231 |
mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) spindle midzone assembly(GO:0051255) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 |
0.1 |
GO:0018206 |
peptidyl-methionine modification(GO:0018206) |
| 0.0 |
0.2 |
GO:0003170 |
heart valve development(GO:0003170) |
| 0.0 |
0.1 |
GO:0001659 |
temperature homeostasis(GO:0001659) |
| 0.0 |
0.1 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
0.1 |
GO:1903755 |
regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 0.0 |
0.1 |
GO:1990173 |
protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.0 |
0.0 |
GO:0045974 |
miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 |
0.1 |
GO:0070232 |
regulation of T cell apoptotic process(GO:0070232) |
| 0.0 |
0.1 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 |
0.1 |
GO:0006515 |
misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 |
0.3 |
GO:0034612 |
response to tumor necrosis factor(GO:0034612) |
| 0.0 |
0.1 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.2 |
GO:0019228 |
neuronal action potential(GO:0019228) |
| 0.0 |
0.1 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 |
0.1 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.0 |
0.1 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.0 |
GO:2000316 |
negative regulation of interferon-gamma secretion(GO:1902714) regulation of T-helper 17 type immune response(GO:2000316) |
| 0.0 |
0.0 |
GO:0010667 |
negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |