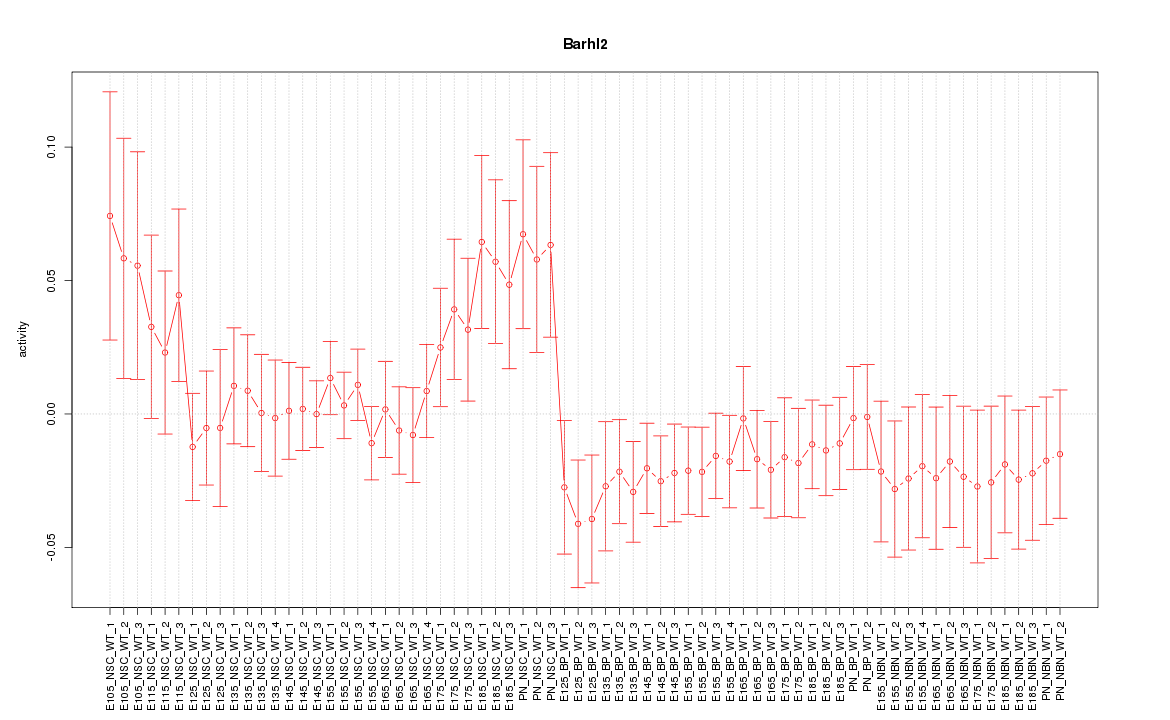
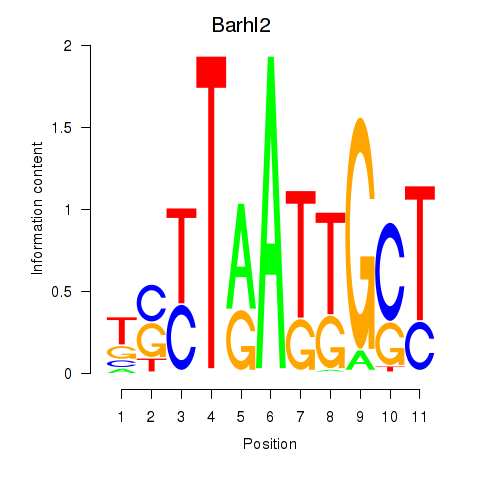
Motif ID: Barhl2
Z-value: 1.036
Transcription factors associated with Barhl2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Barhl2 | ENSMUSG00000034384.10 | Barhl2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Barhl2 | mm10_v2_chr5_-_106458440_106458470 | -0.45 | 8.1e-05 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 15.0 | GO:0035622 | intrahepatic bile duct development(GO:0035622) renal vesicle induction(GO:0072034) |
| 4.3 | 21.4 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 3.9 | 15.8 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 3.8 | 11.5 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) regulation of eosinophil activation(GO:1902566) |
| 3.6 | 14.5 | GO:1903463 | mitotic cell cycle phase(GO:0098763) regulation of mitotic cell cycle DNA replication(GO:1903463) |
| 3.4 | 10.3 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 2.8 | 8.5 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 2.7 | 8.2 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 2.7 | 8.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 2.1 | 38.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 2.0 | 8.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.8 | 5.4 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.5 | 6.0 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 1.4 | 6.8 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 1.3 | 4.0 | GO:0007521 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 1.3 | 9.2 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 1.3 | 5.0 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 1.1 | 14.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 1.1 | 3.3 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.1 | 4.3 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 1.0 | 3.1 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.9 | 6.5 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.9 | 7.3 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.9 | 6.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 | 6.6 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) mitotic DNA replication checkpoint(GO:0033314) |
| 0.8 | 7.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.6 | 10.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.6 | 3.2 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.6 | 1.8 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.6 | 4.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.6 | 1.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.5 | 9.7 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.5 | 4.8 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 4.7 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.5 | 6.6 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.5 | 3.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.4 | 1.3 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.4 | 5.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 | 6.5 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.4 | 1.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 0.3 | 3.0 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.3 | 5.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 5.0 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.3 | 2.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.3 | 3.7 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.3 | 1.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.3 | 9.4 | GO:0009395 | phospholipid catabolic process(GO:0009395) |
| 0.2 | 7.0 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 5.6 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.7 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 1.4 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 1.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 3.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 0.8 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.1 | 2.2 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.6 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 | 9.0 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.1 | 0.9 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.1 | 1.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 1.2 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.1 | 3.7 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.1 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 3.1 | GO:0001541 | ovarian follicle development(GO:0001541) |
| 0.0 | 0.5 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 2.3 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 1.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 5.5 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 4.8 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 5.1 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.4 | GO:0016226 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 8.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.0 | 29.9 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 2.1 | 16.5 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 1.2 | 7.2 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 1.2 | 7.0 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 1.1 | 6.6 | GO:0000796 | condensin complex(GO:0000796) |
| 1.1 | 5.4 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.9 | 3.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.9 | 7.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.8 | 3.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.7 | 5.5 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.6 | 2.2 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.5 | 7.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 2.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 5.6 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.4 | 1.2 | GO:0000802 | transverse filament(GO:0000802) |
| 0.2 | 8.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 2.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.2 | 3.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 3.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.2 | 6.5 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 15.3 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 1.4 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.1 | 4.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 8.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 10.7 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.1 | 16.4 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.1 | 9.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 15.0 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 6.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 13.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 4.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 1.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 4.5 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 2.6 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 5.2 | GO:0005925 | focal adhesion(GO:0005925) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.0 | 9.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 2.9 | 11.5 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 2.1 | 10.3 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.7 | 8.5 | GO:0005113 | patched binding(GO:0005113) |
| 1.7 | 6.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.5 | 9.2 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.2 | 3.5 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 1.1 | 12.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 1.1 | 3.3 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 1.1 | 6.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 1.0 | 6.0 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.8 | 6.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.7 | 7.3 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.7 | 7.2 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.5 | 4.2 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.5 | 9.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.4 | 5.6 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.3 | 40.3 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.3 | 5.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.3 | 1.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.3 | 2.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 9.3 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 1.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.3 | 0.8 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.2 | 5.0 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.2 | 1.4 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 3.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 2.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 19.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 3.0 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 8.2 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 3.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.1 | 7.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 1.8 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 11.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 8.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 5.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.2 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.5 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 10.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 8.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 2.8 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 14.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 1.4 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 35.5 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.0 | 2.2 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.0 | 0.9 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 0.2 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 5.7 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 1.9 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 4.4 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 2.8 | GO:0008234 | cysteine-type peptidase activity(GO:0008234) |
| 0.0 | 1.2 | GO:0047485 | protein N-terminus binding(GO:0047485) |