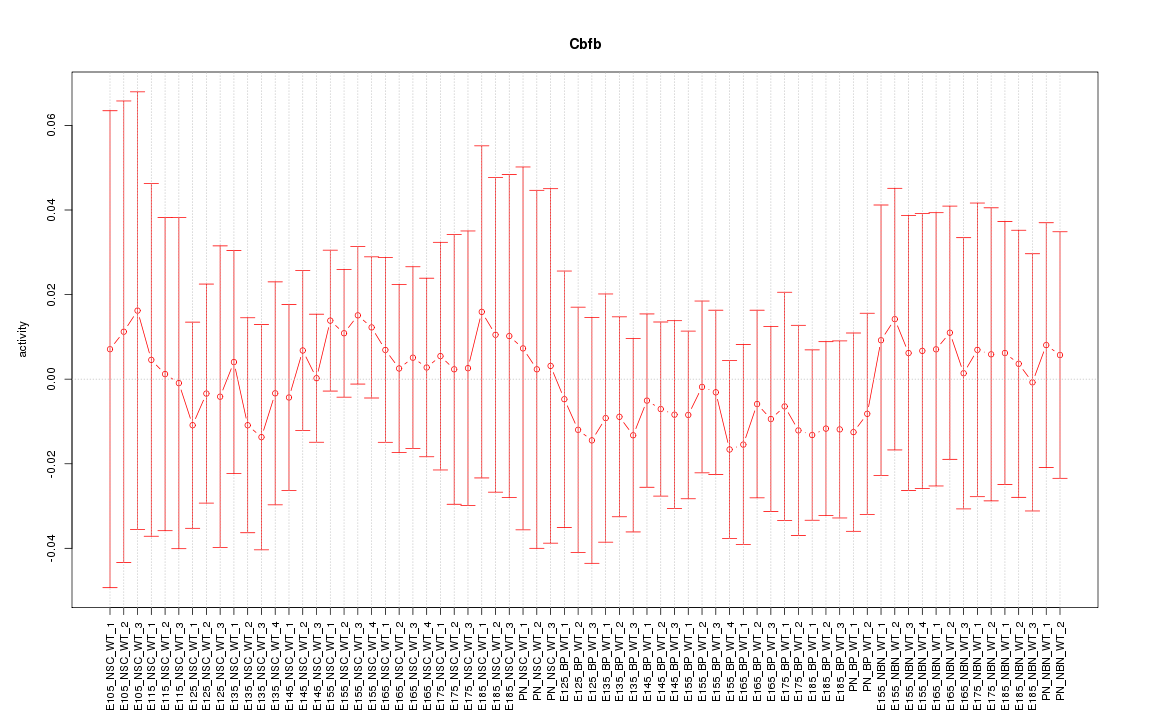
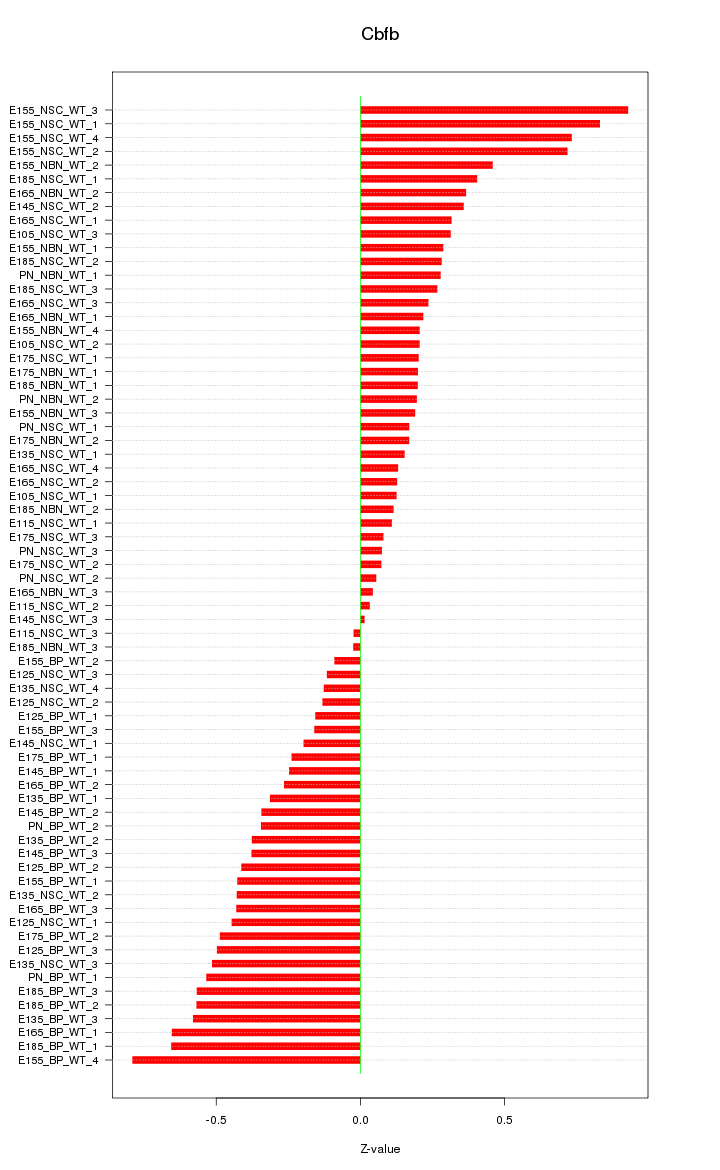
Motif ID: Cbfb
Z-value: 0.372
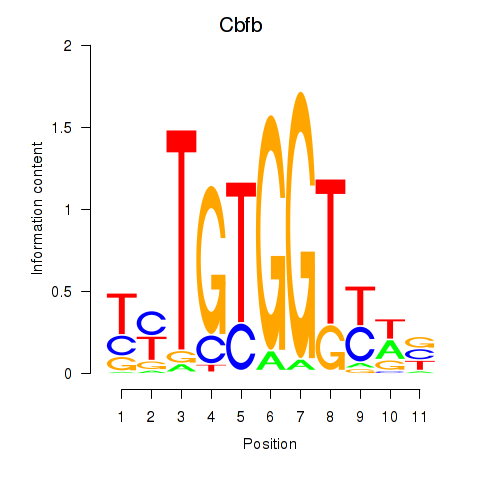
Transcription factors associated with Cbfb:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cbfb | ENSMUSG00000031885.7 | Cbfb |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Cbfb | mm10_v2_chr8_+_105170668_105170686 | 0.18 | 1.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.4 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.2 | 1.1 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.4 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.5 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.4 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.8 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.0 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 2.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) arginine binding(GO:0034618) |
| 0.0 | 1.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) exoribonuclease activity, producing 5'-phosphomonoesters(GO:0016896) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |