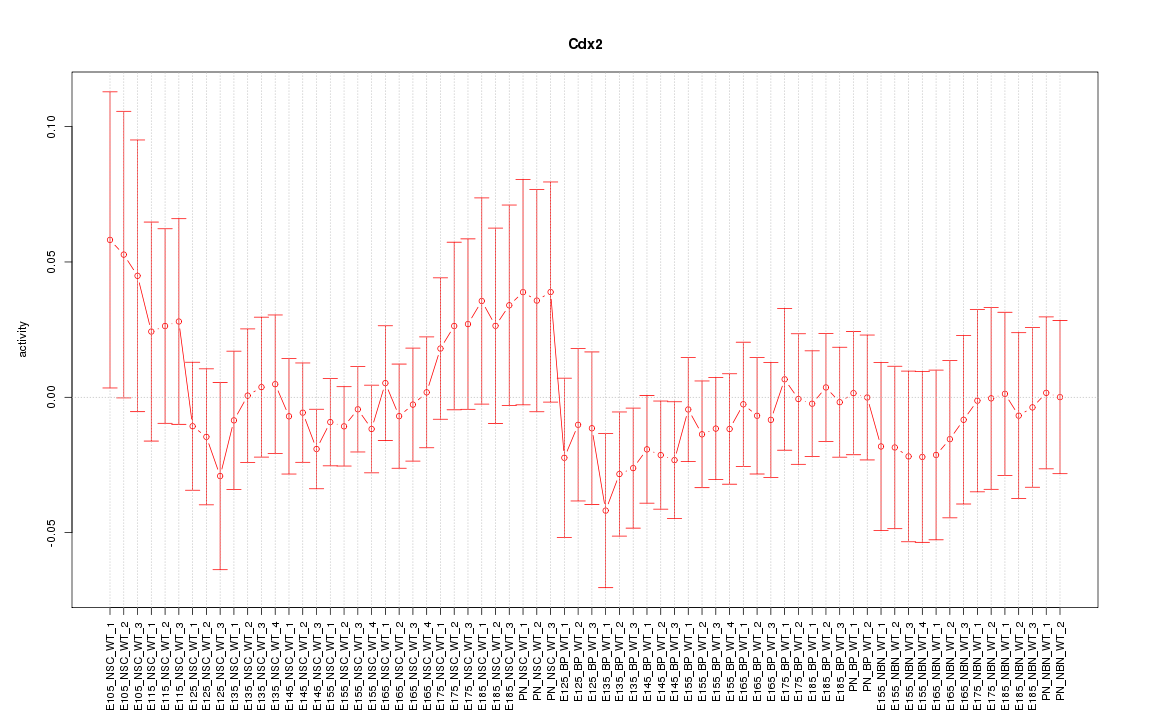
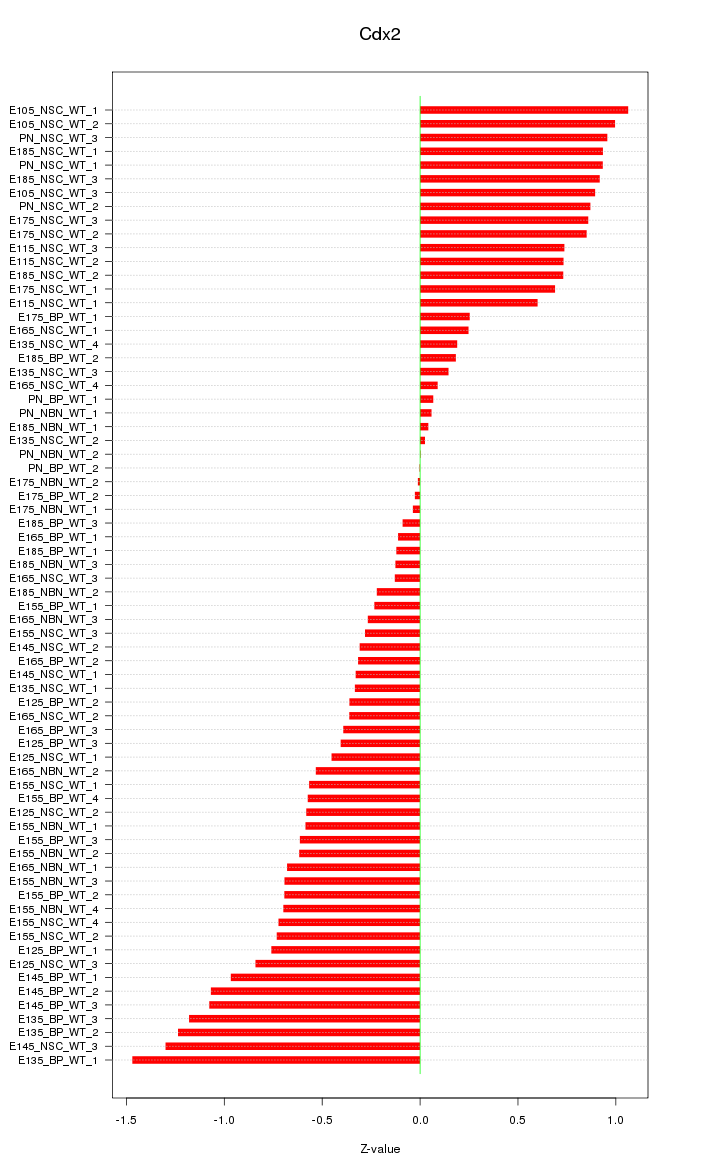
Motif ID: Cdx2
Z-value: 0.649
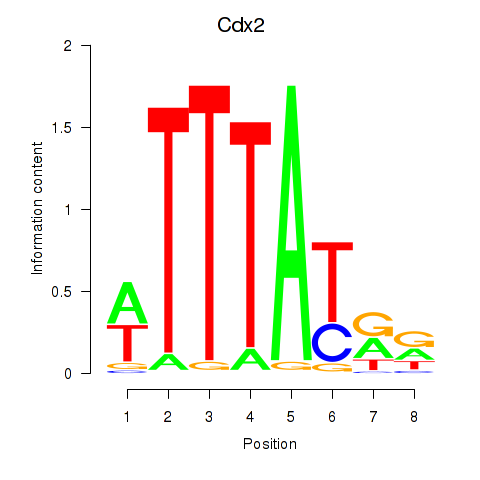
Transcription factors associated with Cdx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Cdx2 | ENSMUSG00000029646.3 | Cdx2 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 1.2 | 5.8 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 1.1 | 3.3 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 1.0 | 3.1 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.0 | 3.1 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.8 | 5.7 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.8 | 3.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.7 | 2.9 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.6 | 3.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.4 | 1.8 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.4 | 3.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.4 | 4.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.4 | 6.7 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.3 | 2.1 | GO:1903887 | motile primary cilium assembly(GO:1903887) |
| 0.3 | 7.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.3 | 2.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 3.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.2 | 4.6 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 1.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 1.9 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 3.5 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 2.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.1 | 0.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 0.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 | 1.2 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 2.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 2.4 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 4.1 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.2 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 0.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0019369 | drug metabolic process(GO:0017144) arachidonic acid metabolic process(GO:0019369) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.5 | 7.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 3.2 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 1.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 3.3 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 4.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 3.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 8.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 4.1 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 3.1 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 10.1 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.2 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 7.8 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 1.4 | 4.1 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 1.3 | 7.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.8 | 3.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.7 | 5.8 | GO:0031432 | titin binding(GO:0031432) |
| 0.7 | 3.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.6 | 3.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.6 | 3.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 5.7 | GO:0008430 | selenium binding(GO:0008430) |
| 0.3 | 1.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 3.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.8 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.2 | 6.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 4.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.2 | 3.1 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 2.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 3.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.9 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 3.1 | GO:0098811 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 3.5 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 1.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 2.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 0.0 | 2.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.8 | GO:0008170 | N-methyltransferase activity(GO:0008170) |