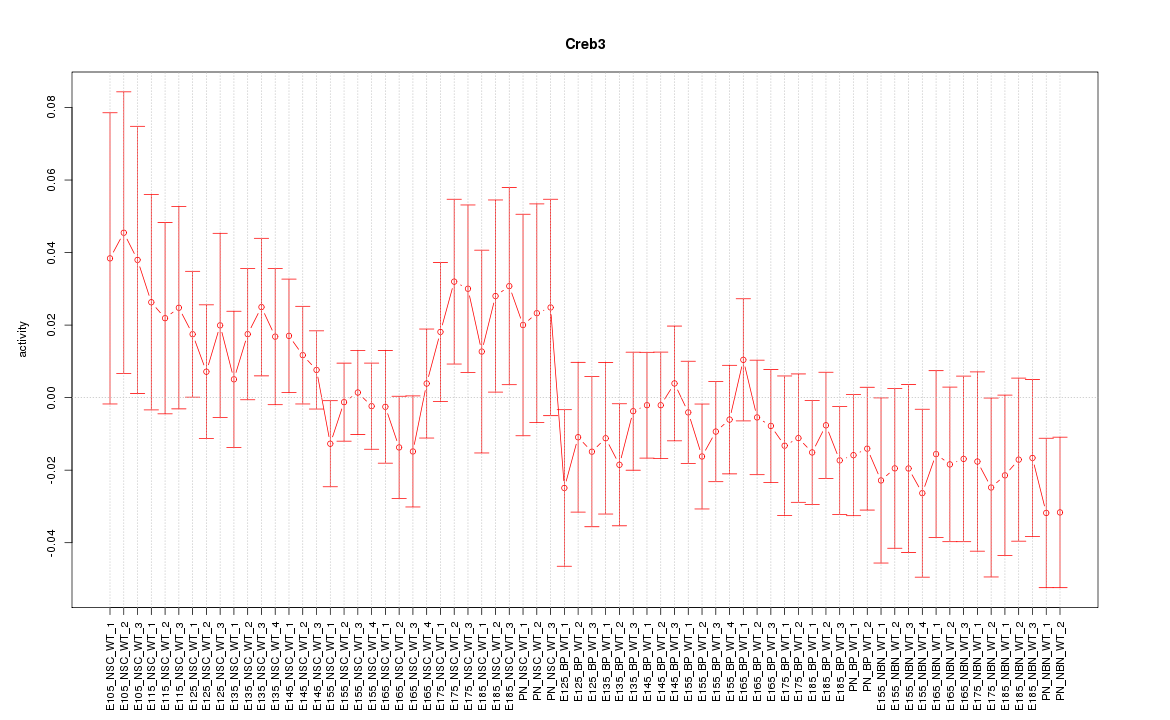
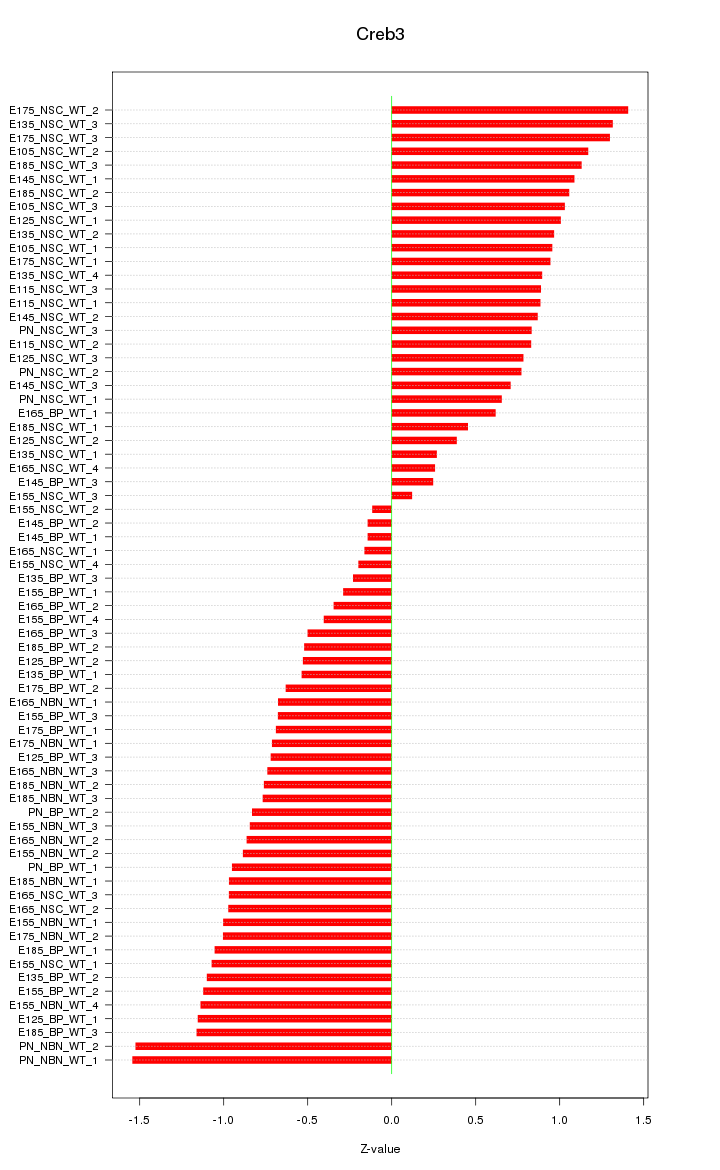
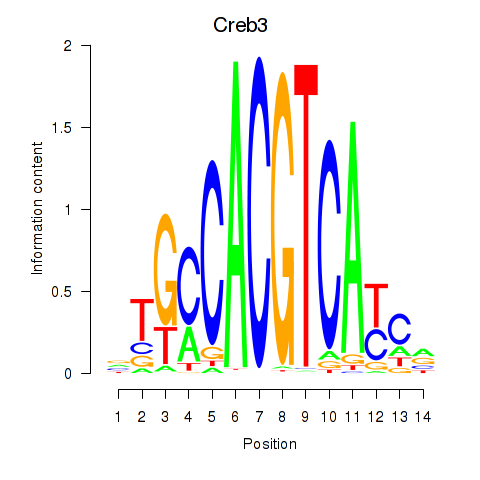
Motif ID: Creb3
Z-value: 0.854
Transcription factors associated with Creb3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Creb3 | ENSMUSG00000028466.9 | Creb3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3 | mm10_v2_chr4_+_43562672_43562947 | -0.73 | 9.6e-13 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 13.7 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 2.7 | 10.8 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 2.3 | 6.8 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.0 | 6.0 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.8 | 12.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.8 | 8.8 | GO:0072429 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 1.7 | 8.3 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 1.7 | 5.0 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.5 | 6.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.3 | 9.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.2 | 5.9 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 1.0 | 6.3 | GO:0007135 | meiosis II(GO:0007135) |
| 0.9 | 2.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.9 | 5.6 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.8 | 5.9 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.8 | 2.4 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.7 | 3.3 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.7 | 2.0 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.6 | 2.5 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.6 | 2.5 | GO:0019287 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.6 | 3.1 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.6 | 1.8 | GO:0050787 | glycolate metabolic process(GO:0009441) enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) glycolate biosynthetic process(GO:0046295) detoxification of mercury ion(GO:0050787) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) glyoxal catabolic process(GO:1903190) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) positive regulation of androgen receptor activity(GO:2000825) |
| 0.6 | 1.7 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.6 | 10.4 | GO:0007530 | sex determination(GO:0007530) |
| 0.6 | 5.0 | GO:1903298 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.5 | 3.6 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.5 | 14.1 | GO:0010664 | negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.5 | 1.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 1.9 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) subthalamic nucleus development(GO:0021763) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) superior vena cava morphogenesis(GO:0060578) |
| 0.5 | 1.9 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.5 | 2.3 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.4 | 1.3 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.4 | 6.0 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.4 | 6.9 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.4 | 2.8 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.4 | 1.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.4 | 1.6 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.4 | 3.1 | GO:1901724 | positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.4 | 1.4 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.3 | 2.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.3 | 1.7 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.3 | 2.6 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.3 | 0.3 | GO:0090298 | negative regulation of mitochondrial DNA replication(GO:0090298) |
| 0.3 | 1.0 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.2 | 2.8 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.2 | 0.7 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 | 1.6 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 | 2.5 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 | 0.9 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.2 | 0.9 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 1.4 | GO:0034397 | telomere localization(GO:0034397) |
| 0.2 | 1.0 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.2 | 2.5 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 0.5 | GO:0010248 | B cell negative selection(GO:0002352) establishment or maintenance of transmembrane electrochemical gradient(GO:0010248) activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.2 | 4.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.1 | 1.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.1 | 4.6 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 3.0 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.1 | 9.2 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.1 | 1.2 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.5 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.1 | 2.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.1 | 2.4 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) regulation of glucose import in response to insulin stimulus(GO:2001273) |
| 0.1 | 1.3 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 5.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.4 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 2.6 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 7.8 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 0.7 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 2.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.1 | 0.4 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.5 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.3 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.5 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 1.3 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 3.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 6.1 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 2.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.1 | 3.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 2.9 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 1.3 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.3 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 1.1 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 3.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.6 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 1.5 | GO:0051272 | positive regulation of cell migration(GO:0030335) positive regulation of cellular component movement(GO:0051272) positive regulation of cell motility(GO:2000147) |
| 0.1 | 1.2 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 1.0 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 | 1.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 3.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.9 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 2.2 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 1.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.9 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 2.7 | GO:0007292 | female gamete generation(GO:0007292) |
| 0.0 | 1.0 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 7.2 | GO:0043010 | camera-type eye development(GO:0043010) |
| 0.0 | 0.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.4 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 1.7 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 3.0 | GO:0048864 | stem cell development(GO:0048864) |
| 0.0 | 0.2 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 2.1 | GO:0001841 | neural tube formation(GO:0001841) |
| 0.0 | 2.5 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.0 | 1.9 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 8.8 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 1.3 | 9.2 | GO:0001940 | male pronucleus(GO:0001940) |
| 1.3 | 3.9 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.6 | 6.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.6 | 2.3 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.5 | 3.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.5 | 4.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.5 | 6.6 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.5 | 3.4 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.5 | 2.3 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 1.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.4 | 1.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.4 | 1.5 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 1.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.3 | 6.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.3 | 0.9 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.3 | 8.3 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 3.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 3.3 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 2.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 9.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 7.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 1.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.2 | 1.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 2.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 1.7 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.7 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.1 | 0.4 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 1.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 6.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.4 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 4.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 2.5 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 3.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 0.4 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 9.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 3.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.9 | GO:0032039 | integrator complex(GO:0032039) |
| 0.1 | 0.5 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 3.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 3.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 3.0 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 3.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 3.1 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 3.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 6.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 2.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.3 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 24.9 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 1.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 8.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 3.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.5 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.0 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 2.3 | 6.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 1.6 | 9.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 1.4 | 5.6 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 1.3 | 3.9 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 1.0 | 4.0 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.9 | 6.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.8 | 4.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.8 | 2.5 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.8 | 2.4 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.8 | 3.9 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.7 | 6.0 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.7 | 2.0 | GO:0070905 | serine binding(GO:0070905) |
| 0.6 | 1.8 | GO:0044388 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) small protein activating enzyme binding(GO:0044388) cupric ion binding(GO:1903135) cuprous ion binding(GO:1903136) glyoxalase (glycolic acid-forming) activity(GO:1990422) |
| 0.6 | 8.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.5 | 1.6 | GO:0034952 | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase activity(GO:0008694) 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2,4-dienoate decarboxylase activity(GO:0018791) bis(4-chlorophenyl)acetate decarboxylase activity(GO:0018792) 3,5-dibromo-4-hydroxybenzoate decarboxylase activity(GO:0018793) 2-hydroxyisobutyrate decarboxylase activity(GO:0018794) 2-hydroxy-2-methyl-1,3-dicarbonate decarboxylase activity(GO:0018795) 2-hydroxyisophthalate decarboxylase activity(GO:0034524) dimethylmalonate decarboxylase activity(GO:0034782) 2,4,4-trimethyl-3-oxopentanoate decarboxylase activity(GO:0034853) 4,4-dimethyl-3-oxopentanoate decarboxylase activity(GO:0034854) 2,3,6-trihydroxyisonicotinate decarboxylase activity(GO:0034879) phenanthrene-4,5-dicarboxylate decarboxylase activity(GO:0034923) pyrrole-2-carboxylate decarboxylase activity(GO:0034941) terephthalate decarboxylase activity(GO:0034947) malonate semialdehyde decarboxylase activity(GO:0034952) 5-amino-4-imidazole carboxylate lyase activity(GO:0043727) 2-keto-4-methylthiobutyrate aminotransferase activity(GO:0043728) 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase activity(GO:0051997) |
| 0.5 | 1.8 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.4 | 4.8 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.4 | 2.6 | GO:0034916 | 4-methyloctanoyl-CoA dehydrogenase activity(GO:0034580) naphthyl-2-methyl-succinyl-CoA dehydrogenase activity(GO:0034845) 2-methylhexanoyl-CoA dehydrogenase activity(GO:0034916) propionyl-CoA dehydrogenase activity(GO:0043820) thiol-driven fumarate reductase activity(GO:0043830) coenzyme F420-dependent 2,4,6-trinitrophenol reductase activity(GO:0052758) coenzyme F420-dependent 2,4,6-trinitrophenol hydride reductase activity(GO:0052759) coenzyme F420-dependent 2,4-dinitrophenol reductase activity(GO:0052760) |
| 0.4 | 9.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.4 | 1.6 | GO:0004096 | catalase activity(GO:0004096) |
| 0.4 | 3.2 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.4 | 1.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.3 | 3.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.3 | 1.0 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 3.3 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 2.8 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.3 | 9.9 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.3 | 10.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.3 | 1.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.3 | 6.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 1.4 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.3 | 3.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.3 | 2.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 1.8 | GO:0070191 | methionine-R-sulfoxide reductase activity(GO:0070191) |
| 0.3 | 2.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.2 | 6.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.2 | 20.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 0.7 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.2 | 3.3 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 4.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.3 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.2 | 1.7 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.2 | 2.9 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 1.9 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.2 | 4.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.2 | 5.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.3 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.1 | 0.9 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 2.1 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 2.8 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 5.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 2.8 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.9 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 4.3 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 13.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.1 | 1.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 8.8 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.1 | 2.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.5 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 2.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 32.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.4 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.9 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.1 | 1.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 2.5 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 1.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.9 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 3.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 4.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 6.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 3.2 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 4.7 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 1.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.3 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 0.1 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.0 | 3.0 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 1.1 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 1.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |