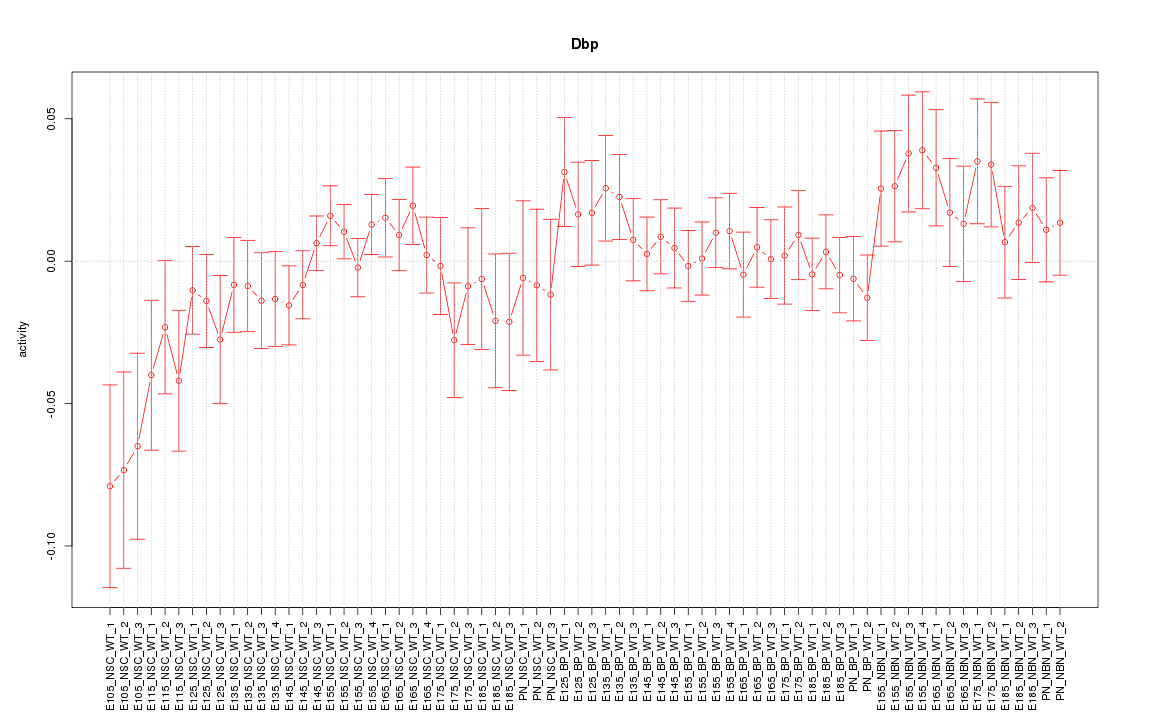
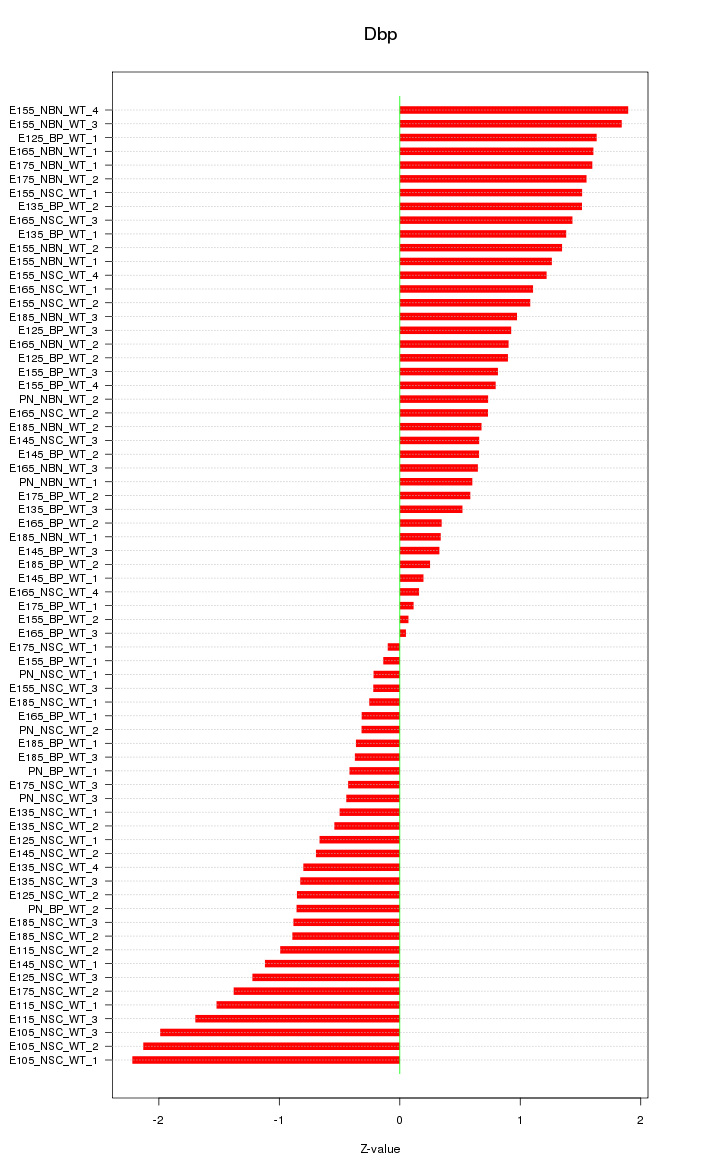
Motif ID: Dbp
Z-value: 1.024
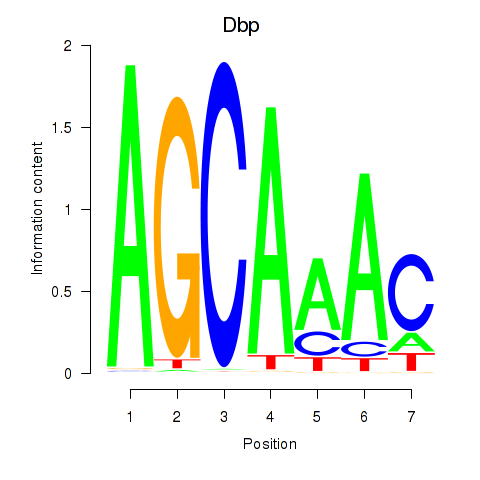
Transcription factors associated with Dbp:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dbp | ENSMUSG00000059824.4 | Dbp |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbp | mm10_v2_chr7_+_45705088_45705292 | 0.37 | 1.5e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.3 | 15.8 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 3.3 | 9.8 | GO:0021852 | pyramidal neuron migration(GO:0021852) |
| 2.8 | 14.1 | GO:2000302 | regulation of synaptic vesicle endocytosis(GO:1900242) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 2.0 | 6.1 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 2.0 | 8.0 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 2.0 | 10.0 | GO:0071205 | regulation of axon diameter(GO:0031133) clustering of voltage-gated potassium channels(GO:0045163) protein localization to juxtaparanode region of axon(GO:0071205) |
| 2.0 | 3.9 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 1.8 | 24.7 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 1.7 | 11.7 | GO:0071361 | cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 1.6 | 4.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.6 | 14.3 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 1.6 | 14.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.5 | 4.6 | GO:0003289 | atrial septum primum morphogenesis(GO:0003289) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.5 | 13.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 1.5 | 4.4 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 1.4 | 12.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 1.4 | 4.1 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 1.4 | 16.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 1.3 | 7.6 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 1.3 | 10.0 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.2 | 3.7 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) regulation of nucleoside transport(GO:0032242) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) |
| 1.2 | 9.9 | GO:0097264 | self proteolysis(GO:0097264) |
| 1.1 | 14.8 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 1.1 | 3.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.0 | 3.9 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 1.0 | 16.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 1.0 | 20.3 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.9 | 5.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.9 | 2.8 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.8 | 11.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.8 | 4.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.8 | 2.3 | GO:0061526 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) musculoskeletal movement, spinal reflex action(GO:0050883) acetylcholine secretion(GO:0061526) |
| 0.7 | 9.7 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.7 | 5.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.6 | 3.8 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.6 | 1.9 | GO:0048686 | regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.6 | 3.1 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.6 | 6.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.6 | 2.4 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.5 | 4.4 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.5 | 5.6 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.5 | 5.5 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.5 | 2.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.5 | 2.4 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.5 | 2.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 6.5 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.4 | 7.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.4 | 14.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 2.5 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.4 | 13.1 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.4 | 1.6 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.4 | 4.8 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.4 | 2.8 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 | 8.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 23.6 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 2.2 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.3 | 1.8 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 | 2.1 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.3 | 5.4 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.3 | 1.4 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.3 | 2.2 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.3 | 1.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 | 2.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.3 | 4.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.3 | 2.5 | GO:1901621 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 9.8 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.2 | 9.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 0.7 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) |
| 0.2 | 4.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.2 | 1.8 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.2 | 8.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 1.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.2 | 2.9 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.2 | 0.6 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.2 | 2.6 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.2 | 3.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 1.5 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.1 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.2 | 4.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.8 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.2 | 2.4 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 4.8 | GO:0030890 | positive regulation of B cell proliferation(GO:0030890) |
| 0.2 | 1.0 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.2 | 1.8 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.2 | 13.4 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.2 | 2.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 1.0 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.2 | 0.8 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.2 | 2.9 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 0.1 | 8.5 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 10.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.5 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.9 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.9 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 0.1 | 1.5 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 2.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 1.8 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.1 | 6.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 9.8 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.1 | 9.3 | GO:0098792 | xenophagy(GO:0098792) |
| 0.1 | 4.8 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 1.0 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 5.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 1.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 2.1 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 5.6 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 2.9 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 | 1.6 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.1 | 5.0 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.1 | 1.1 | GO:0060043 | regulation of cardiac muscle cell proliferation(GO:0060043) |
| 0.1 | 1.3 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 6.7 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 2.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 5.6 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 6.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 1.3 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 1.0 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 2.0 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 1.2 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 1.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 1.9 | GO:0000086 | G2/M transition of mitotic cell cycle(GO:0000086) |
| 0.0 | 1.0 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.2 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.7 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 3.2 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0031673 | H zone(GO:0031673) |
| 1.3 | 33.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 1.2 | 11.0 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 1.2 | 24.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.9 | 5.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.9 | 15.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.8 | 15.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 3.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.7 | 8.0 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.6 | 3.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.6 | 6.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.6 | 2.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.6 | 1.8 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.6 | 1.2 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.6 | 5.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 11.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.5 | 10.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 3.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.5 | 3.9 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.5 | 2.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.3 | 10.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 26.7 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.3 | 16.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 2.4 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.3 | 3.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.2 | 4.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 2.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.2 | 2.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 44.9 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 35.4 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.2 | 3.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.2 | 1.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.8 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.2 | 4.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 9.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 9.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 13.3 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.1 | 0.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 6.1 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 2.5 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.1 | 1.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 3.9 | GO:0031252 | cell leading edge(GO:0031252) |
| 0.1 | 6.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 6.0 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 21.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 1.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 12.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 6.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 1.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 13.3 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 2.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 5.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 1.8 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 9.6 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 5.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.0 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 1.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 11.7 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 3.5 | 24.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 2.7 | 8.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 1.8 | 14.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.7 | 13.5 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 1.4 | 11.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.4 | 11.0 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.4 | 4.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 1.3 | 3.8 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 1.3 | 6.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.9 | 2.8 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.9 | 12.8 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.9 | 2.6 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.9 | 15.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.8 | 15.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.8 | 3.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.8 | 14.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.7 | 2.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.6 | 3.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.6 | 6.1 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.5 | 5.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 26.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 3.7 | GO:0032795 | G-protein coupled adenosine receptor activity(GO:0001609) heterotrimeric G-protein binding(GO:0032795) |
| 0.5 | 1.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) ErbB-3 class receptor binding(GO:0043125) |
| 0.5 | 2.4 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.5 | 1.9 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.5 | 2.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.4 | 2.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 2.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.3 | 2.4 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.3 | 5.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 4.8 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.3 | 2.9 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.3 | 1.1 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 1.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 1.6 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 1.8 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.3 | 2.4 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.3 | 3.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 0.8 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.3 | 5.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 3.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 8.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 1.0 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.2 | 11.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 10.0 | GO:0043621 | protein self-association(GO:0043621) |
| 0.2 | 5.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 3.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.2 | 3.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 13.5 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.2 | 7.4 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 4.8 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 1.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.1 | 2.0 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 8.1 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 2.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 2.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 1.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.8 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 1.0 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 3.6 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.1 | 19.2 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.1 | 1.6 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.6 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.1 | 8.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 8.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.1 | 3.8 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.1 | 2.2 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.0 | 2.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 7.3 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 1.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 1.0 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 18.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 3.2 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 5.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 7.9 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.5 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 2.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0042288 | GTPase activating protein binding(GO:0032794) MHC class I protein binding(GO:0042288) |