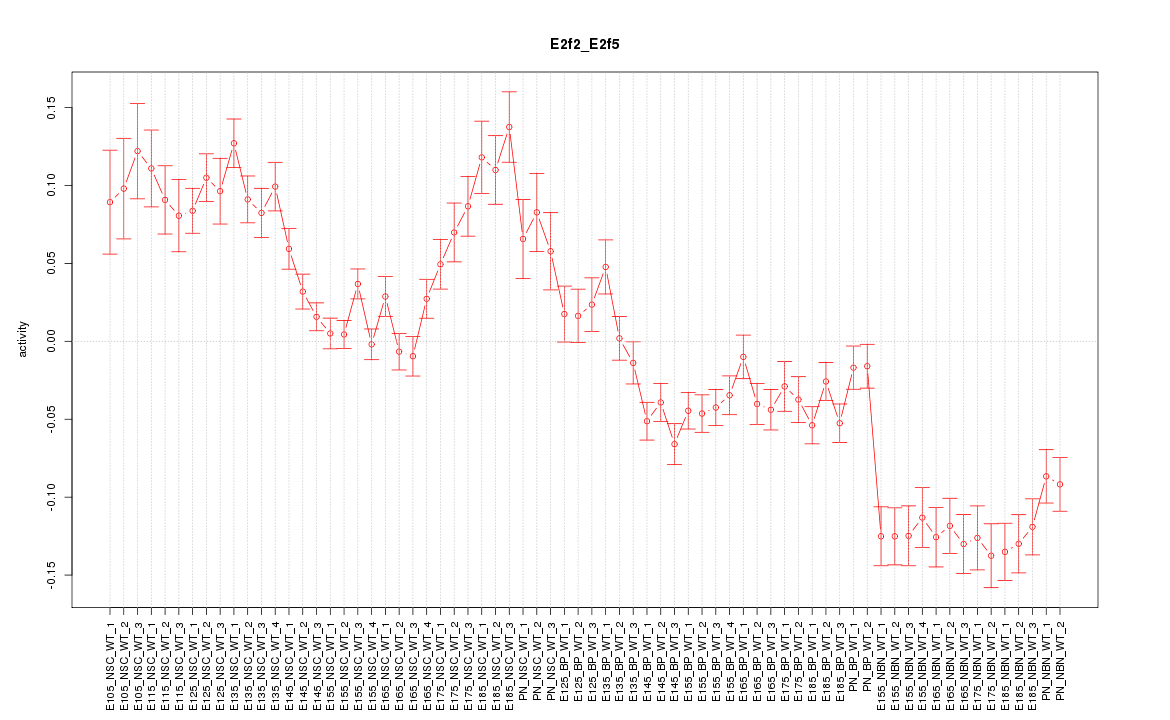
Motif ID: E2f2_E2f5
Z-value: 4.385
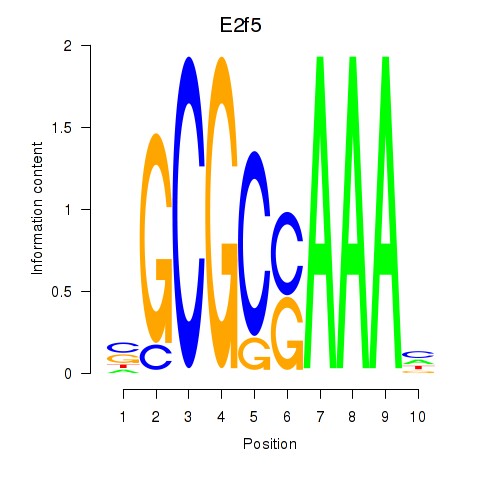
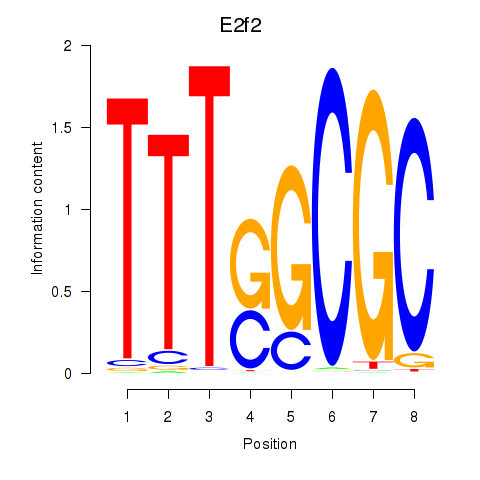
Transcription factors associated with E2f2_E2f5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| E2f2 | ENSMUSG00000018983.9 | E2f2 |
| E2f5 | ENSMUSG00000027552.8 | E2f5 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f2 | mm10_v2_chr4_+_136172367_136172395 | 0.97 | 1.6e-44 | Click! |
| E2f5 | mm10_v2_chr3_+_14578609_14578687 | 0.90 | 1.2e-26 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 40.1 | 120.2 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 25.5 | 76.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 20.4 | 142.8 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 15.7 | 125.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 13.2 | 171.5 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 13.0 | 38.9 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 12.2 | 48.8 | GO:0046655 | glycine biosynthetic process(GO:0006545) folic acid metabolic process(GO:0046655) |
| 11.5 | 45.8 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 11.0 | 263.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 11.0 | 153.5 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 10.6 | 42.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 10.6 | 105.9 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 10.2 | 81.7 | GO:0033504 | floor plate development(GO:0033504) |
| 9.3 | 83.8 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 9.1 | 72.5 | GO:0019985 | translesion synthesis(GO:0019985) |
| 8.9 | 44.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 8.5 | 25.6 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 8.4 | 33.5 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 8.2 | 41.0 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 7.6 | 68.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 7.5 | 30.1 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 7.5 | 45.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 7.0 | 28.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) |
| 6.1 | 18.3 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 6.1 | 48.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 5.9 | 59.0 | GO:0015884 | positive regulation of platelet activation(GO:0010572) folic acid transport(GO:0015884) |
| 5.7 | 22.6 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) negative regulation of respiratory burst(GO:0060268) |
| 5.5 | 33.2 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 4.9 | 39.0 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 4.7 | 28.5 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 4.6 | 27.6 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 4.5 | 18.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 3.6 | 18.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 3.6 | 17.8 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 3.5 | 3.5 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 3.5 | 17.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 3.3 | 10.0 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 3.2 | 19.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 3.2 | 15.9 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 3.0 | 15.0 | GO:0042148 | strand invasion(GO:0042148) |
| 2.9 | 17.5 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 2.9 | 171.9 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 2.9 | 17.1 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 2.8 | 5.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 2.7 | 10.8 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 2.7 | 8.0 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 2.4 | 7.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 2.3 | 11.3 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 2.2 | 13.0 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 2.1 | 33.8 | GO:0006298 | mismatch repair(GO:0006298) |
| 2.1 | 93.3 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 2.0 | 8.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.9 | 25.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 1.9 | 7.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 1.9 | 7.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.9 | 5.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 1.9 | 7.4 | GO:2001032 | regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 1.8 | 18.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 1.8 | 9.0 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 1.7 | 5.2 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.7 | 5.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 1.7 | 5.0 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) |
| 1.6 | 14.7 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 1.6 | 4.9 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 1.6 | 11.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.6 | 21.8 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 1.5 | 15.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 1.4 | 5.7 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.2 | 4.8 | GO:0061010 | gall bladder development(GO:0061010) |
| 1.2 | 4.7 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 1.2 | 5.8 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.1 | 10.0 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 1.0 | 10.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 1.0 | 11.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 1.0 | 4.1 | GO:1904008 | response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 1.0 | 13.0 | GO:0030238 | male sex determination(GO:0030238) |
| 1.0 | 9.8 | GO:0003093 | regulation of glomerular filtration(GO:0003093) positive regulation of integrin-mediated signaling pathway(GO:2001046) |
| 1.0 | 2.9 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.9 | 3.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.9 | 14.9 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.9 | 6.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.9 | 21.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.9 | 2.7 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.8 | 13.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.8 | 18.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.8 | 2.4 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.8 | 7.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.8 | 2.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.8 | 3.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.7 | 2.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.7 | 2.1 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.7 | 6.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.7 | 45.6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.7 | 15.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.7 | 4.8 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.7 | 2.7 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.7 | 25.9 | GO:0006289 | nucleotide-excision repair(GO:0006289) |
| 0.7 | 2.6 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.6 | 10.9 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) reciprocal DNA recombination(GO:0035825) |
| 0.6 | 36.8 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.6 | 13.5 | GO:0034728 | nucleosome assembly(GO:0006334) nucleosome organization(GO:0034728) |
| 0.6 | 12.5 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.5 | 1.1 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.5 | 4.5 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 6.5 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.5 | 7.9 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.5 | 3.5 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.5 | 7.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.5 | 3.6 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.4 | 2.7 | GO:1904251 | regulation of bile acid metabolic process(GO:1904251) |
| 0.4 | 17.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.4 | 0.8 | GO:2000015 | regulation of determination of dorsal identity(GO:2000015) |
| 0.4 | 64.1 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.4 | 8.9 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.4 | 5.7 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.3 | 5.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.3 | 1.7 | GO:0006983 | ER overload response(GO:0006983) |
| 0.3 | 2.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 3.3 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.3 | 0.6 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.3 | 3.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.3 | 7.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.2 | 3.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 | 7.3 | GO:0060135 | maternal process involved in female pregnancy(GO:0060135) |
| 0.2 | 5.1 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.2 | 17.0 | GO:0045860 | positive regulation of protein kinase activity(GO:0045860) |
| 0.2 | 2.4 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.2 | 3.8 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.2 | 6.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.2 | 0.5 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) noradrenergic neuron differentiation(GO:0003357) |
| 0.2 | 4.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.2 | 1.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 10.0 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.2 | 1.0 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.2 | 3.9 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.2 | 1.0 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.2 | 10.9 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.2 | 1.7 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.2 | 3.2 | GO:0032094 | response to food(GO:0032094) |
| 0.2 | 20.4 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.2 | 0.8 | GO:0071557 | notochord morphogenesis(GO:0048570) histone H3-K27 demethylation(GO:0071557) |
| 0.2 | 10.9 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 1.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 2.2 | GO:0000291 | nuclear-transcribed mRNA catabolic process, exonucleolytic(GO:0000291) |
| 0.1 | 8.4 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.1 | 2.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 8.5 | GO:0007569 | cell aging(GO:0007569) |
| 0.1 | 6.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.1 | 3.0 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 1.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 4.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.6 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.1 | 1.3 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 0.3 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.1 | 3.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 5.8 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 0.8 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.1 | 1.0 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 1.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.5 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.9 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.6 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 2.0 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.7 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 2.0 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 1.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.4 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.5 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.4 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 23.8 | 118.9 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 18.8 | 282.7 | GO:0042555 | MCM complex(GO:0042555) |
| 16.2 | 16.2 | GO:0000811 | GINS complex(GO:0000811) |
| 14.8 | 44.4 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 11.7 | 58.5 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 8.7 | 43.3 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 7.1 | 42.3 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 6.5 | 19.6 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 5.6 | 44.6 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 4.7 | 47.2 | GO:0000800 | lateral element(GO:0000800) |
| 4.1 | 218.0 | GO:0005657 | replication fork(GO:0005657) |
| 3.5 | 35.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 2.6 | 15.6 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 2.3 | 13.8 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 2.0 | 10.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.8 | 14.4 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 1.7 | 5.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 1.6 | 95.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 1.6 | 10.9 | GO:0070652 | HAUS complex(GO:0070652) |
| 1.5 | 6.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 1.5 | 13.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 1.4 | 13.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 1.3 | 28.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 1.2 | 18.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.2 | 68.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 1.1 | 5.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 1.1 | 5.5 | GO:0005683 | U7 snRNP(GO:0005683) |
| 1.1 | 6.6 | GO:0070187 | telosome(GO:0070187) |
| 1.1 | 3.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.8 | 10.9 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.8 | 6.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.8 | 2.4 | GO:0001740 | Barr body(GO:0001740) |
| 0.8 | 26.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.7 | 2.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.7 | 71.4 | GO:0005814 | centriole(GO:0005814) |
| 0.7 | 307.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.6 | 4.7 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.6 | 39.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.6 | 4.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.6 | 1.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.6 | 2.9 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.6 | 8.4 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.6 | 2.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.5 | 6.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.5 | 28.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.5 | 3.7 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) telomeric heterochromatin(GO:0031933) |
| 0.5 | 3.5 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 11.1 | GO:0002102 | podosome(GO:0002102) |
| 0.5 | 5.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.4 | 8.9 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 7.1 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.4 | 0.8 | GO:0000801 | central element(GO:0000801) |
| 0.4 | 32.3 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.4 | 2.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.4 | 21.7 | GO:0000922 | spindle pole(GO:0000922) |
| 0.4 | 13.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.4 | 35.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.3 | 2.8 | GO:0000243 | commitment complex(GO:0000243) |
| 0.3 | 36.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.3 | 8.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.3 | 19.9 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 6.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.3 | 0.8 | GO:0090543 | Flemming body(GO:0090543) |
| 0.3 | 11.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 2.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.2 | 7.9 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.2 | 10.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.2 | 1.0 | GO:0001652 | granular component(GO:0001652) |
| 0.2 | 1.7 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.2 | 70.6 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 0.4 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 1.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 5.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 12.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.2 | 2.8 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 14.7 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 1.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 34.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 8.1 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 1.2 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 1.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 56.0 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 0.8 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 0.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 29.6 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 413.5 | GO:0005634 | nucleus(GO:0005634) |
| 0.1 | 2.5 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.1 | 5.1 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 7.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.5 | GO:0035770 | ribonucleoprotein granule(GO:0035770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 46.7 | 140.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 23.8 | 118.9 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 20.9 | 125.6 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 19.4 | 155.0 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 16.3 | 48.8 | GO:0051870 | methotrexate binding(GO:0051870) |
| 15.1 | 60.4 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 8.9 | 62.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 8.5 | 34.0 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 7.0 | 27.8 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 6.6 | 59.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 6.5 | 6.5 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 6.4 | 115.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 5.1 | 35.9 | GO:0003896 | DNA primase activity(GO:0003896) |
| 4.9 | 93.8 | GO:0035173 | histone kinase activity(GO:0035173) |
| 4.1 | 44.6 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 3.9 | 19.3 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 3.7 | 11.2 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 3.6 | 96.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 3.6 | 17.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 3.6 | 24.9 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 3.3 | 10.0 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 3.3 | 13.0 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 2.8 | 17.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 2.8 | 8.4 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 2.8 | 33.0 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 2.7 | 162.6 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 2.6 | 38.9 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 2.5 | 12.4 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 2.4 | 29.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 2.4 | 7.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 2.0 | 18.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 1.9 | 76.6 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 1.8 | 5.5 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 1.8 | 22.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 1.8 | 132.3 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 1.7 | 10.0 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.4 | 17.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 1.4 | 5.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.4 | 11.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 1.4 | 248.8 | GO:0001047 | core promoter binding(GO:0001047) |
| 1.3 | 6.6 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 1.3 | 28.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 1.2 | 3.6 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 1.1 | 7.9 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 1.1 | 1.1 | GO:0016751 | S-succinyltransferase activity(GO:0016751) |
| 1.0 | 68.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 1.0 | 40.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.9 | 14.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.8 | 5.0 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.7 | 14.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.7 | 40.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.7 | 2.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.7 | 15.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.7 | 2.8 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.7 | 4.8 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.7 | 11.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.6 | 3.2 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.6 | 1.3 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.6 | 5.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.5 | 64.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.5 | 2.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.5 | 2.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.4 | 2.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.4 | 1.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.4 | 16.5 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.4 | 3.7 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.4 | 4.9 | GO:0048038 | quinone binding(GO:0048038) |
| 0.4 | 47.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.3 | 21.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.3 | 8.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 5.8 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.3 | 2.4 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 86.0 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.3 | 15.1 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 0.9 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.3 | 3.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 15.3 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.3 | 2.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.3 | 2.9 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 3.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 44.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.2 | 5.6 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 21.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 6.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.2 | 0.8 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 4.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.2 | 3.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.2 | 0.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.2 | 3.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.2 | 7.1 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.2 | 6.7 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.2 | 5.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.2 | 1.2 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.2 | 6.0 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) |
| 0.2 | 106.9 | GO:0044212 | transcription regulatory region DNA binding(GO:0044212) |
| 0.2 | 2.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.7 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 16.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 10.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.4 | GO:0043747 | protein-N-terminal asparagine amidohydrolase activity(GO:0008418) UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity(GO:0008759) iprodione amidohydrolase activity(GO:0018748) (3,5-dichlorophenylurea)acetate amidohydrolase activity(GO:0018749) 4'-(2-hydroxyisopropyl)phenylurea amidohydrolase activity(GO:0034571) didemethylisoproturon amidohydrolase activity(GO:0034573) N-isopropylacetanilide amidohydrolase activity(GO:0034576) N-cyclohexylformamide amidohydrolase activity(GO:0034781) isonicotinic acid hydrazide hydrolase activity(GO:0034876) cis-aconitamide amidase activity(GO:0034882) gamma-N-formylaminovinylacetate hydrolase activity(GO:0034885) N2-acetyl-L-lysine deacetylase activity(GO:0043747) O-succinylbenzoate synthase activity(GO:0043748) indoleacetamide hydrolase activity(GO:0043864) N-acetylcitrulline deacetylase activity(GO:0043909) N-acetylgalactosamine-6-phosphate deacetylase activity(GO:0047419) diacetylchitobiose deacetylase activity(GO:0052773) chitooligosaccharide deacetylase activity(GO:0052790) |
| 0.1 | 0.4 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 2.0 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 2.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 9.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.4 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 3.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 19.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.6 | GO:0016462 | pyrophosphatase activity(GO:0016462) |
| 0.1 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.1 | 7.6 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 5.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 0.7 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 1.0 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 3.3 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 4.8 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 1.0 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 3.1 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.3 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |