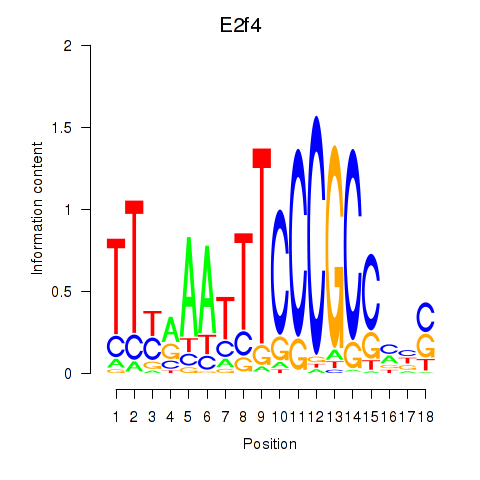
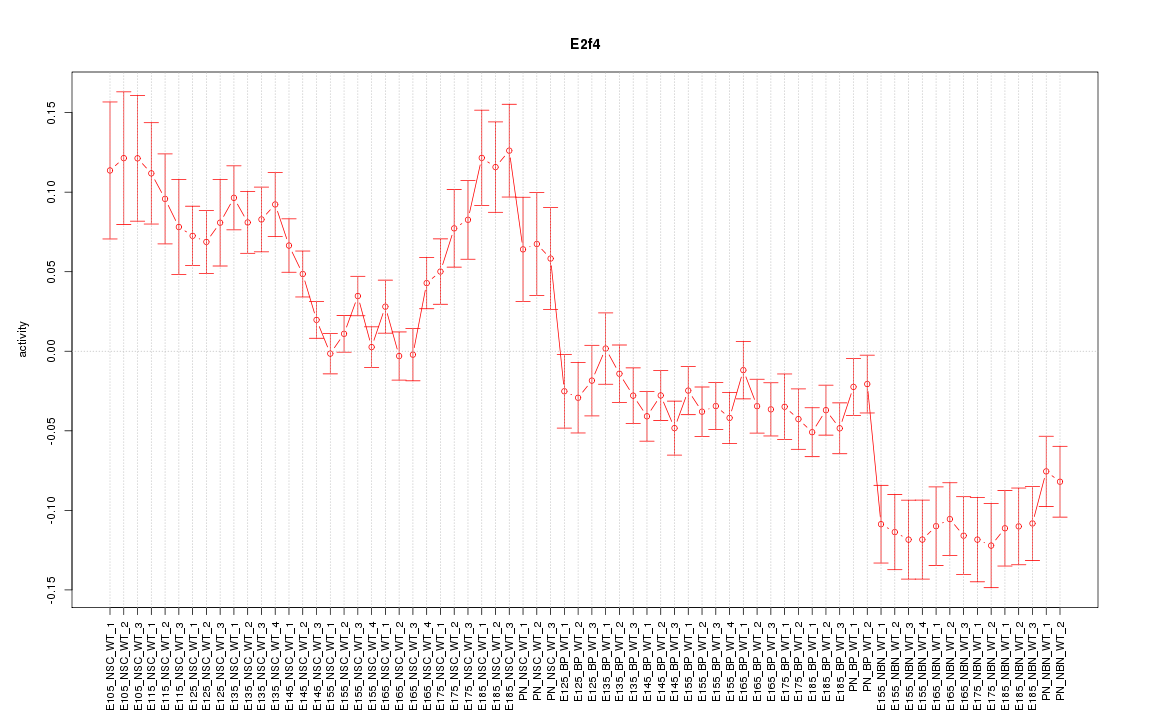
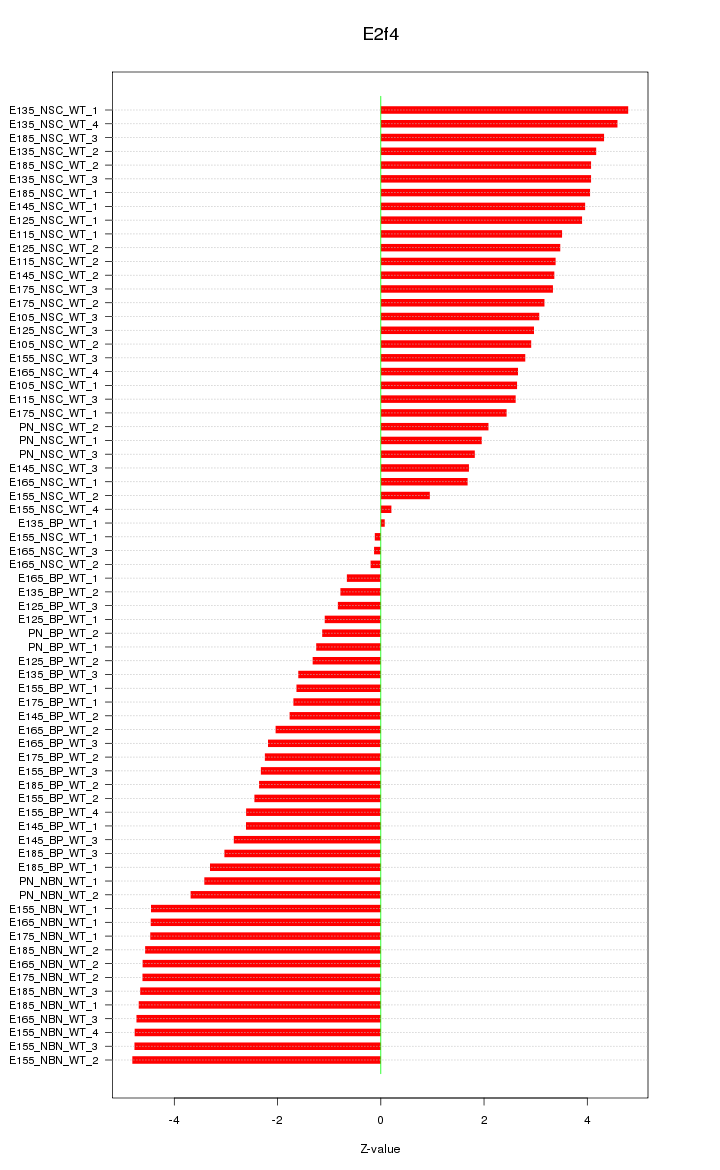
| 18.7 |
56.2 |
GO:0071921 |
establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 17.8 |
71.4 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 16.5 |
49.4 |
GO:0000087 |
mitotic M phase(GO:0000087) |
| 12.0 |
35.9 |
GO:0003219 |
atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 11.8 |
35.5 |
GO:0072076 |
hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 11.1 |
33.3 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 9.9 |
29.7 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 8.2 |
24.6 |
GO:0010911 |
regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 6.3 |
44.3 |
GO:2000042 |
negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 5.6 |
79.0 |
GO:0070986 |
left/right axis specification(GO:0070986) |
| 5.6 |
22.4 |
GO:0000707 |
meiotic DNA recombinase assembly(GO:0000707) |
| 4.4 |
31.1 |
GO:0000320 |
re-entry into mitotic cell cycle(GO:0000320) |
| 3.8 |
18.9 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 3.7 |
33.5 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 3.6 |
28.8 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 3.5 |
6.9 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 3.3 |
82.2 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 3.2 |
19.4 |
GO:0007144 |
female meiosis I(GO:0007144) |
| 3.1 |
15.7 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 2.9 |
8.6 |
GO:0000710 |
meiotic mismatch repair(GO:0000710) |
| 2.6 |
84.6 |
GO:0008608 |
attachment of spindle microtubules to kinetochore(GO:0008608) |
| 2.5 |
55.2 |
GO:0008340 |
determination of adult lifespan(GO:0008340) |
| 2.5 |
17.5 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 2.4 |
9.8 |
GO:1902990 |
mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 2.3 |
9.3 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 2.2 |
17.9 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 1.9 |
24.7 |
GO:0051984 |
positive regulation of chromosome segregation(GO:0051984) |
| 1.8 |
7.2 |
GO:0035878 |
nail development(GO:0035878) |
| 1.7 |
5.2 |
GO:0060821 |
inactivation of X chromosome by DNA methylation(GO:0060821) |
| 1.7 |
13.3 |
GO:0010804 |
negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 1.7 |
36.4 |
GO:0051310 |
metaphase plate congression(GO:0051310) |
| 1.6 |
4.9 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 1.6 |
31.8 |
GO:1900153 |
regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 1.5 |
9.0 |
GO:0060623 |
regulation of chromosome condensation(GO:0060623) |
| 1.5 |
11.8 |
GO:0007016 |
cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 1.5 |
5.9 |
GO:1904008 |
response to monosodium glutamate(GO:1904008) cellular response to monosodium glutamate(GO:1904009) |
| 1.5 |
33.6 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 1.5 |
129.1 |
GO:0007052 |
mitotic spindle organization(GO:0007052) |
| 1.4 |
13.6 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 1.3 |
16.1 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 1.1 |
3.3 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 1.1 |
6.4 |
GO:0046909 |
intermembrane transport(GO:0046909) |
| 1.0 |
18.8 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 1.0 |
54.6 |
GO:0007051 |
spindle organization(GO:0007051) |
| 1.0 |
6.8 |
GO:0070561 |
vitamin D receptor signaling pathway(GO:0070561) |
| 0.9 |
5.6 |
GO:0043353 |
enucleate erythrocyte differentiation(GO:0043353) |
| 0.9 |
37.0 |
GO:0006284 |
base-excision repair(GO:0006284) |
| 0.9 |
13.9 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.9 |
12.8 |
GO:0042640 |
anagen(GO:0042640) |
| 0.9 |
3.5 |
GO:0046125 |
pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.9 |
27.9 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 0.8 |
5.0 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.7 |
33.1 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.7 |
7.2 |
GO:0051608 |
organic cation transport(GO:0015695) histamine transport(GO:0051608) |
| 0.7 |
18.5 |
GO:0006910 |
phagocytosis, recognition(GO:0006910) |
| 0.7 |
2.1 |
GO:0043987 |
histone-serine phosphorylation(GO:0035404) histone H3-S10 phosphorylation(GO:0043987) |
| 0.7 |
6.9 |
GO:0036123 |
histone H3-K9 dimethylation(GO:0036123) |
| 0.6 |
5.1 |
GO:0090219 |
negative regulation of lipid kinase activity(GO:0090219) |
| 0.6 |
5.8 |
GO:0043568 |
positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.6 |
1.7 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 |
6.6 |
GO:0009143 |
nucleoside triphosphate catabolic process(GO:0009143) |
| 0.5 |
6.8 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.5 |
4.6 |
GO:0000012 |
single strand break repair(GO:0000012) |
| 0.4 |
12.9 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 |
6.8 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.4 |
2.8 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.4 |
4.8 |
GO:0033210 |
leptin-mediated signaling pathway(GO:0033210) |
| 0.4 |
1.5 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.3 |
1.0 |
GO:0060161 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.3 |
1.6 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.3 |
2.9 |
GO:0035881 |
amacrine cell differentiation(GO:0035881) |
| 0.3 |
1.3 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.3 |
1.3 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 |
0.9 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.3 |
5.8 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.3 |
3.7 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.2 |
11.3 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.2 |
69.5 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.2 |
0.6 |
GO:0014043 |
negative regulation of neuron maturation(GO:0014043) |
| 0.2 |
0.5 |
GO:0007227 |
signal transduction downstream of smoothened(GO:0007227) positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 |
1.4 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.2 |
9.2 |
GO:0035690 |
cellular response to drug(GO:0035690) |
| 0.1 |
1.8 |
GO:0006120 |
mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 |
11.8 |
GO:0000956 |
nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.1 |
8.7 |
GO:0006334 |
nucleosome assembly(GO:0006334) |
| 0.1 |
1.6 |
GO:0042790 |
transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 |
5.4 |
GO:1902593 |
protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) nuclear import(GO:0051170) single-organism nuclear import(GO:1902593) |
| 0.1 |
4.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.1 |
1.1 |
GO:0061050 |
regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.1 |
0.3 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.1 |
4.4 |
GO:0015914 |
phospholipid transport(GO:0015914) |
| 0.1 |
0.8 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
2.5 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.1 |
0.3 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.1 |
5.1 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.0 |
0.5 |
GO:0097067 |
cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 |
5.3 |
GO:0019221 |
cytokine-mediated signaling pathway(GO:0019221) |
| 0.0 |
3.5 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.0 |
22.9 |
GO:0007049 |
cell cycle(GO:0007049) |
| 0.0 |
12.8 |
GO:0008284 |
positive regulation of cell proliferation(GO:0008284) |
| 0.0 |
1.7 |
GO:0098792 |
xenophagy(GO:0098792) |
| 0.0 |
0.5 |
GO:0010212 |
response to ionizing radiation(GO:0010212) |