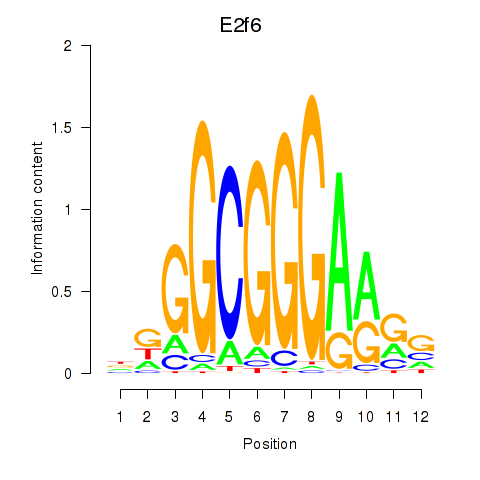
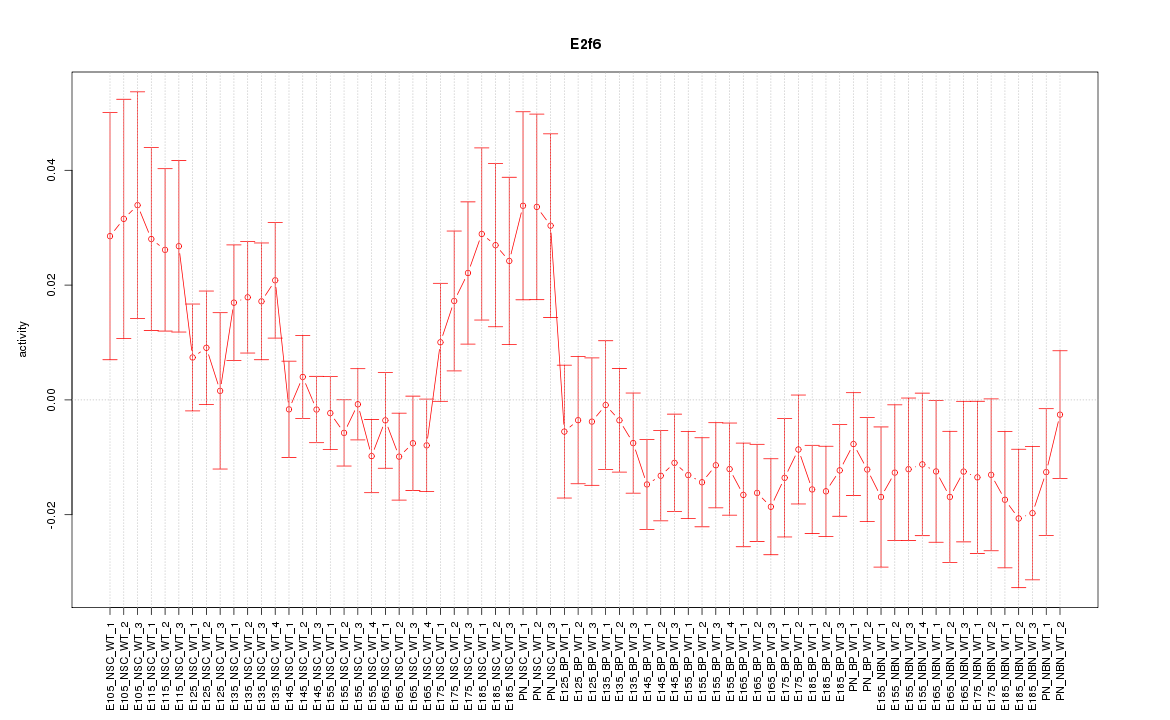
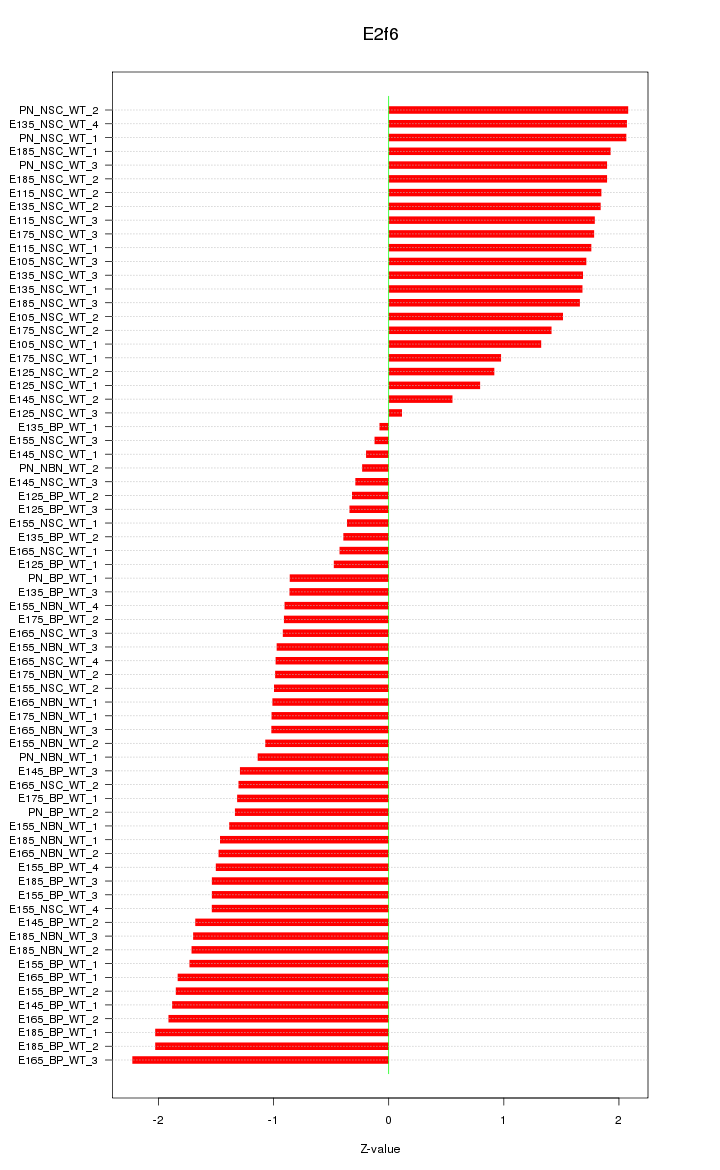
| 13.3 |
39.9 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 7.3 |
21.8 |
GO:0030421 |
defecation(GO:0030421) |
| 6.8 |
13.6 |
GO:0072180 |
mesonephric duct development(GO:0072177) mesonephric duct morphogenesis(GO:0072180) |
| 5.5 |
16.5 |
GO:0021529 |
spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 5.2 |
62.2 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 4.4 |
17.5 |
GO:2000525 |
regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 4.4 |
4.4 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 3.8 |
11.5 |
GO:0060129 |
thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 3.8 |
7.6 |
GO:0072197 |
renal vesicle induction(GO:0072034) ureter morphogenesis(GO:0072197) |
| 3.5 |
3.5 |
GO:0021593 |
rhombomere morphogenesis(GO:0021593) |
| 3.4 |
13.6 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 3.3 |
9.8 |
GO:0021759 |
globus pallidus development(GO:0021759) |
| 3.2 |
6.5 |
GO:0061144 |
alveolar secondary septum development(GO:0061144) |
| 3.2 |
9.6 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 3.1 |
12.5 |
GO:0015888 |
thiamine transport(GO:0015888) |
| 2.9 |
8.6 |
GO:0003221 |
right ventricular cardiac muscle tissue morphogenesis(GO:0003221) ventricular compact myocardium morphogenesis(GO:0003223) |
| 2.8 |
13.9 |
GO:0006348 |
chromatin silencing at telomere(GO:0006348) |
| 2.7 |
13.7 |
GO:1900170 |
negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 2.7 |
8.1 |
GO:1900158 |
negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 2.7 |
8.1 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 2.6 |
5.3 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 2.6 |
7.8 |
GO:0061743 |
motor learning(GO:0061743) |
| 2.6 |
10.3 |
GO:0003360 |
brainstem development(GO:0003360) |
| 2.6 |
7.7 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 2.5 |
12.5 |
GO:0030916 |
otic vesicle formation(GO:0030916) |
| 2.4 |
7.3 |
GO:0006566 |
threonine metabolic process(GO:0006566) |
| 2.4 |
2.4 |
GO:0072554 |
blood vessel lumenization(GO:0072554) |
| 2.3 |
18.8 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 2.3 |
14.1 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 2.3 |
9.3 |
GO:0009957 |
epidermal cell fate specification(GO:0009957) |
| 2.1 |
2.1 |
GO:0070094 |
positive regulation of glucagon secretion(GO:0070094) |
| 2.1 |
10.4 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 2.1 |
2.1 |
GO:0003256 |
regulation of transcription from RNA polymerase II promoter involved in myocardial precursor cell differentiation(GO:0003256) |
| 2.0 |
6.1 |
GO:0060364 |
frontal suture morphogenesis(GO:0060364) |
| 2.0 |
6.1 |
GO:0045659 |
eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 2.0 |
30.0 |
GO:0007530 |
sex determination(GO:0007530) |
| 2.0 |
31.7 |
GO:0009263 |
deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.9 |
5.8 |
GO:0035907 |
dorsal aorta development(GO:0035907) dorsal aorta morphogenesis(GO:0035912) |
| 1.9 |
5.7 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 1.8 |
7.3 |
GO:0003150 |
muscular septum morphogenesis(GO:0003150) |
| 1.8 |
7.3 |
GO:0061113 |
pancreas morphogenesis(GO:0061113) |
| 1.8 |
1.8 |
GO:0098743 |
cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 1.8 |
7.2 |
GO:0046084 |
adenine metabolic process(GO:0046083) adenine biosynthetic process(GO:0046084) |
| 1.8 |
12.4 |
GO:0033567 |
DNA replication, Okazaki fragment processing(GO:0033567) |
| 1.8 |
8.8 |
GO:0001705 |
ectoderm formation(GO:0001705) |
| 1.7 |
5.2 |
GO:0001866 |
NK T cell proliferation(GO:0001866) |
| 1.7 |
8.6 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 1.7 |
5.1 |
GO:0060447 |
bud outgrowth involved in lung branching(GO:0060447) |
| 1.6 |
4.7 |
GO:0006269 |
DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.6 |
4.7 |
GO:1905005 |
regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905005) |
| 1.5 |
4.6 |
GO:0045409 |
negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 1.5 |
12.3 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 1.5 |
6.0 |
GO:0071394 |
cellular response to testosterone stimulus(GO:0071394) |
| 1.4 |
4.3 |
GO:0060244 |
negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.4 |
8.6 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.4 |
5.7 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) succinate transport(GO:0015744) citrate transport(GO:0015746) |
| 1.4 |
1.4 |
GO:0007352 |
zygotic specification of dorsal/ventral axis(GO:0007352) |
| 1.3 |
4.0 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.3 |
40.0 |
GO:0006270 |
DNA replication initiation(GO:0006270) |
| 1.3 |
3.8 |
GO:0007161 |
calcium-independent cell-matrix adhesion(GO:0007161) |
| 1.3 |
8.9 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 1.3 |
5.0 |
GO:0010288 |
response to lead ion(GO:0010288) |
| 1.2 |
6.2 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 1.2 |
7.4 |
GO:0006344 |
maintenance of chromatin silencing(GO:0006344) |
| 1.2 |
3.7 |
GO:0048133 |
germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 1.2 |
9.8 |
GO:0001675 |
acrosome assembly(GO:0001675) |
| 1.2 |
12.1 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 1.2 |
6.0 |
GO:1903553 |
positive regulation of extracellular exosome assembly(GO:1903553) |
| 1.2 |
5.9 |
GO:0007089 |
traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.2 |
4.7 |
GO:0014722 |
regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 1.2 |
12.9 |
GO:0010216 |
maintenance of DNA methylation(GO:0010216) |
| 1.1 |
3.4 |
GO:0002184 |
cytoplasmic translational termination(GO:0002184) |
| 1.1 |
4.5 |
GO:0045578 |
negative regulation of B cell differentiation(GO:0045578) |
| 1.1 |
7.8 |
GO:0006297 |
nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 1.1 |
4.4 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.1 |
3.3 |
GO:0086017 |
renin secretion into blood stream(GO:0002001) cell communication by chemical coupling(GO:0010643) Purkinje myocyte action potential(GO:0086017) regulation of renin secretion into blood stream(GO:1900133) |
| 1.1 |
3.2 |
GO:1901552 |
positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 1.1 |
6.5 |
GO:0051461 |
regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 1.1 |
1.1 |
GO:0046822 |
regulation of nucleocytoplasmic transport(GO:0046822) |
| 1.1 |
3.2 |
GO:0007084 |
mitotic nuclear envelope reassembly(GO:0007084) |
| 1.1 |
4.3 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 1.1 |
3.2 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 1.1 |
11.6 |
GO:0031659 |
positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 1.1 |
28.4 |
GO:0043153 |
entrainment of circadian clock by photoperiod(GO:0043153) |
| 1.0 |
2.1 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 1.0 |
4.2 |
GO:0070829 |
heterochromatin maintenance(GO:0070829) |
| 1.0 |
4.1 |
GO:1900145 |
regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 1.0 |
3.0 |
GO:0051295 |
establishment of meiotic spindle localization(GO:0051295) |
| 1.0 |
4.0 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.0 |
1.0 |
GO:0046101 |
hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 1.0 |
3.9 |
GO:1904426 |
positive regulation of GTP binding(GO:1904426) regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 1.0 |
9.7 |
GO:0060235 |
lens induction in camera-type eye(GO:0060235) |
| 1.0 |
8.8 |
GO:0048096 |
chromatin-mediated maintenance of transcription(GO:0048096) |
| 1.0 |
4.8 |
GO:0021797 |
forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.0 |
2.9 |
GO:0015014 |
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.9 |
11.3 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.9 |
2.8 |
GO:2000297 |
negative regulation of synapse maturation(GO:2000297) |
| 0.9 |
12.0 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.9 |
2.7 |
GO:0010957 |
negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.9 |
2.7 |
GO:1901675 |
negative regulation of histone H3-K27 acetylation(GO:1901675) |
| 0.9 |
4.5 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 0.9 |
7.3 |
GO:0033327 |
Leydig cell differentiation(GO:0033327) |
| 0.9 |
2.7 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.9 |
10.8 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.9 |
3.5 |
GO:0051305 |
chromosome movement towards spindle pole(GO:0051305) |
| 0.9 |
5.3 |
GO:0003433 |
chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.9 |
3.4 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.8 |
20.4 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.8 |
4.2 |
GO:0006369 |
termination of RNA polymerase II transcription(GO:0006369) |
| 0.8 |
7.5 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.8 |
2.5 |
GO:0018214 |
peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.8 |
4.8 |
GO:0060040 |
retinal bipolar neuron differentiation(GO:0060040) |
| 0.8 |
6.3 |
GO:0045603 |
positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.8 |
3.9 |
GO:0060836 |
lymphatic endothelial cell differentiation(GO:0060836) |
| 0.8 |
3.9 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.8 |
3.1 |
GO:0006526 |
arginine biosynthetic process(GO:0006526) |
| 0.8 |
7.7 |
GO:0002089 |
lens morphogenesis in camera-type eye(GO:0002089) |
| 0.8 |
4.6 |
GO:0070836 |
caveola assembly(GO:0070836) |
| 0.8 |
3.8 |
GO:0019852 |
L-ascorbic acid metabolic process(GO:0019852) |
| 0.8 |
3.8 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.8 |
2.3 |
GO:0090271 |
positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.7 |
2.2 |
GO:0030167 |
proteoglycan catabolic process(GO:0030167) |
| 0.7 |
2.9 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.7 |
2.2 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.7 |
2.1 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.7 |
3.5 |
GO:0038018 |
Wnt receptor catabolic process(GO:0038018) |
| 0.7 |
7.0 |
GO:0043011 |
myeloid dendritic cell differentiation(GO:0043011) |
| 0.7 |
4.1 |
GO:0060510 |
Type II pneumocyte differentiation(GO:0060510) |
| 0.7 |
0.7 |
GO:0003149 |
membranous septum morphogenesis(GO:0003149) |
| 0.7 |
8.8 |
GO:0046643 |
regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.7 |
3.4 |
GO:0010032 |
meiotic chromosome condensation(GO:0010032) |
| 0.7 |
1.3 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.7 |
13.8 |
GO:0040034 |
regulation of development, heterochronic(GO:0040034) |
| 0.7 |
3.3 |
GO:0010961 |
cellular magnesium ion homeostasis(GO:0010961) |
| 0.6 |
10.4 |
GO:0061029 |
eyelid development in camera-type eye(GO:0061029) |
| 0.6 |
0.6 |
GO:0033128 |
negative regulation of histone phosphorylation(GO:0033128) |
| 0.6 |
3.9 |
GO:1904948 |
midbrain dopaminergic neuron differentiation(GO:1904948) |
| 0.6 |
1.9 |
GO:0030208 |
dermatan sulfate biosynthetic process(GO:0030208) |
| 0.6 |
1.9 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.6 |
6.3 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 0.6 |
3.8 |
GO:0043586 |
tongue development(GO:0043586) |
| 0.6 |
1.2 |
GO:0039530 |
MDA-5 signaling pathway(GO:0039530) |
| 0.6 |
2.5 |
GO:0006268 |
DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 |
6.1 |
GO:0070940 |
dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.6 |
0.6 |
GO:0003418 |
growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.6 |
4.2 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.6 |
3.5 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.6 |
8.2 |
GO:0038092 |
nodal signaling pathway(GO:0038092) |
| 0.6 |
1.7 |
GO:2001269 |
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.6 |
11.9 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.6 |
1.1 |
GO:0070425 |
negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) |
| 0.6 |
3.9 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.6 |
5.0 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.6 |
1.7 |
GO:1902915 |
negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.6 |
0.6 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.5 |
3.8 |
GO:1903689 |
regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.5 |
1.1 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.5 |
5.4 |
GO:0033234 |
negative regulation of protein sumoylation(GO:0033234) |
| 0.5 |
3.2 |
GO:0060736 |
prostate gland growth(GO:0060736) |
| 0.5 |
2.1 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.5 |
7.3 |
GO:0050434 |
positive regulation of viral transcription(GO:0050434) |
| 0.5 |
9.2 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.5 |
2.6 |
GO:0001561 |
fatty acid alpha-oxidation(GO:0001561) |
| 0.5 |
1.5 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.5 |
1.5 |
GO:0021957 |
corticospinal tract morphogenesis(GO:0021957) |
| 0.5 |
2.9 |
GO:0032825 |
positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.5 |
22.5 |
GO:2000134 |
negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.5 |
13.6 |
GO:0046677 |
response to antibiotic(GO:0046677) |
| 0.5 |
6.3 |
GO:0001709 |
cell fate determination(GO:0001709) |
| 0.5 |
1.4 |
GO:1901165 |
positive regulation of trophoblast cell migration(GO:1901165) |
| 0.5 |
4.2 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.5 |
3.2 |
GO:0097237 |
cellular response to toxic substance(GO:0097237) |
| 0.5 |
6.4 |
GO:0038063 |
collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.4 |
1.8 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.4 |
4.9 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.4 |
3.1 |
GO:0034379 |
very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.4 |
2.2 |
GO:0006741 |
NADP biosynthetic process(GO:0006741) |
| 0.4 |
2.6 |
GO:0036066 |
protein O-linked fucosylation(GO:0036066) |
| 0.4 |
7.0 |
GO:0021800 |
cerebral cortex tangential migration(GO:0021800) |
| 0.4 |
4.8 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 |
3.0 |
GO:0018158 |
protein oxidation(GO:0018158) |
| 0.4 |
2.2 |
GO:0060430 |
lung saccule development(GO:0060430) |
| 0.4 |
0.9 |
GO:0042045 |
epithelial fluid transport(GO:0042045) |
| 0.4 |
0.9 |
GO:1904251 |
regulation of bile acid metabolic process(GO:1904251) |
| 0.4 |
0.8 |
GO:2000018 |
regulation of male gonad development(GO:2000018) positive regulation of male gonad development(GO:2000020) |
| 0.4 |
1.7 |
GO:0072235 |
distal convoluted tubule development(GO:0072025) DCT cell differentiation(GO:0072069) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) metanephric DCT cell differentiation(GO:0072240) |
| 0.4 |
3.3 |
GO:0031547 |
brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.4 |
1.2 |
GO:0019079 |
viral genome replication(GO:0019079) |
| 0.4 |
6.6 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.4 |
1.2 |
GO:1905154 |
negative regulation of tumor necrosis factor secretion(GO:1904468) negative regulation of membrane invagination(GO:1905154) |
| 0.4 |
3.2 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.4 |
1.6 |
GO:0000415 |
negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.4 |
2.8 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.4 |
2.3 |
GO:0030644 |
cellular chloride ion homeostasis(GO:0030644) |
| 0.4 |
1.2 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.4 |
1.5 |
GO:1903546 |
protein localization to photoreceptor outer segment(GO:1903546) |
| 0.4 |
1.5 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.4 |
1.5 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.4 |
2.9 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.4 |
4.7 |
GO:0050774 |
negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 |
1.8 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.4 |
1.4 |
GO:0043654 |
recognition of apoptotic cell(GO:0043654) |
| 0.4 |
0.7 |
GO:0042636 |
negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 0.4 |
1.8 |
GO:0042711 |
maternal behavior(GO:0042711) |
| 0.4 |
10.7 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 |
1.8 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.4 |
1.1 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 |
2.8 |
GO:0007076 |
mitotic chromosome condensation(GO:0007076) |
| 0.3 |
3.1 |
GO:0061299 |
retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 |
1.0 |
GO:1904154 |
protein localization to endoplasmic reticulum exit site(GO:0070973) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.3 |
1.4 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
7.2 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.3 |
3.4 |
GO:0035988 |
chondrocyte proliferation(GO:0035988) |
| 0.3 |
1.7 |
GO:0061687 |
detoxification of inorganic compound(GO:0061687) |
| 0.3 |
4.1 |
GO:0090136 |
epithelial cell-cell adhesion(GO:0090136) |
| 0.3 |
3.7 |
GO:0036297 |
interstrand cross-link repair(GO:0036297) |
| 0.3 |
0.7 |
GO:0030997 |
regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 |
0.7 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.3 |
1.7 |
GO:0001840 |
neural plate development(GO:0001840) |
| 0.3 |
1.3 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.3 |
4.6 |
GO:0051292 |
nuclear pore complex assembly(GO:0051292) |
| 0.3 |
1.0 |
GO:0006116 |
NADH oxidation(GO:0006116) |
| 0.3 |
1.9 |
GO:0007296 |
vitellogenesis(GO:0007296) |
| 0.3 |
0.6 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.3 |
0.6 |
GO:0010533 |
regulation of activation of Janus kinase activity(GO:0010533) regulation of activation of JAK2 kinase activity(GO:0010534) |
| 0.3 |
2.5 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 |
3.7 |
GO:0034377 |
plasma lipoprotein particle assembly(GO:0034377) |
| 0.3 |
1.2 |
GO:0016259 |
selenocysteine metabolic process(GO:0016259) |
| 0.3 |
2.1 |
GO:0008343 |
adult feeding behavior(GO:0008343) |
| 0.3 |
0.9 |
GO:0015772 |
disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.3 |
2.0 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.3 |
1.4 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.3 |
6.4 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.3 |
0.3 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.3 |
1.1 |
GO:0046602 |
regulation of mitotic centrosome separation(GO:0046602) |
| 0.3 |
0.8 |
GO:0015910 |
peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.3 |
3.8 |
GO:0051602 |
response to electrical stimulus(GO:0051602) |
| 0.3 |
1.4 |
GO:0016266 |
O-glycan processing(GO:0016266) |
| 0.3 |
7.5 |
GO:0048844 |
artery morphogenesis(GO:0048844) |
| 0.3 |
1.3 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.3 |
1.9 |
GO:0042135 |
neurotransmitter catabolic process(GO:0042135) |
| 0.3 |
2.9 |
GO:0031936 |
negative regulation of chromatin silencing(GO:0031936) |
| 0.3 |
8.1 |
GO:0007588 |
excretion(GO:0007588) |
| 0.3 |
0.5 |
GO:0033184 |
positive regulation of histone ubiquitination(GO:0033184) |
| 0.3 |
2.6 |
GO:0045899 |
positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.3 |
0.8 |
GO:0038180 |
nerve growth factor signaling pathway(GO:0038180) |
| 0.3 |
2.3 |
GO:0031339 |
negative regulation of vesicle fusion(GO:0031339) |
| 0.2 |
2.2 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.2 |
2.0 |
GO:0006362 |
transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.2 |
2.7 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 |
0.7 |
GO:0001828 |
inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.2 |
0.7 |
GO:0046368 |
GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 |
1.2 |
GO:1903300 |
negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 |
0.9 |
GO:1903265 |
positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.2 |
1.1 |
GO:0070934 |
CRD-mediated mRNA stabilization(GO:0070934) |
| 0.2 |
0.9 |
GO:2000144 |
positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.2 |
0.7 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 |
2.5 |
GO:0010225 |
response to UV-C(GO:0010225) |
| 0.2 |
1.8 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.2 |
2.0 |
GO:1903960 |
negative regulation of anion transmembrane transport(GO:1903960) |
| 0.2 |
1.3 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.2 |
0.9 |
GO:1902774 |
late endosome to lysosome transport(GO:1902774) |
| 0.2 |
5.3 |
GO:0045070 |
positive regulation of viral genome replication(GO:0045070) |
| 0.2 |
1.1 |
GO:0019236 |
response to pheromone(GO:0019236) |
| 0.2 |
13.8 |
GO:0007229 |
integrin-mediated signaling pathway(GO:0007229) |
| 0.2 |
0.6 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.2 |
1.3 |
GO:0001996 |
regulation of systemic arterial blood pressure by norepinephrine-epinephrine(GO:0001993) positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of heart rate by epinephrine(GO:0003065) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.2 |
0.8 |
GO:2001274 |
negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 |
0.8 |
GO:0097298 |
regulation of nucleus size(GO:0097298) |
| 0.2 |
1.7 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 |
3.4 |
GO:0071577 |
zinc II ion transmembrane transport(GO:0071577) |
| 0.2 |
1.6 |
GO:0001569 |
patterning of blood vessels(GO:0001569) |
| 0.2 |
7.5 |
GO:0035307 |
positive regulation of protein dephosphorylation(GO:0035307) |
| 0.2 |
1.4 |
GO:0098535 |
de novo centriole assembly(GO:0098535) |
| 0.2 |
0.8 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.2 |
4.3 |
GO:0006379 |
mRNA cleavage(GO:0006379) |
| 0.2 |
1.2 |
GO:0061087 |
positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.2 |
1.6 |
GO:0006398 |
mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 |
1.9 |
GO:0006490 |
oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.2 |
3.6 |
GO:0070208 |
protein heterotrimerization(GO:0070208) |
| 0.2 |
0.7 |
GO:0060459 |
left lung development(GO:0060459) endodermal digestive tract morphogenesis(GO:0061031) |
| 0.2 |
4.8 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.2 |
2.3 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.2 |
1.9 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.2 |
0.3 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.2 |
1.4 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.2 |
1.9 |
GO:0043518 |
negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) |
| 0.2 |
1.4 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.2 |
0.3 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.2 |
0.2 |
GO:0030838 |
positive regulation of actin filament polymerization(GO:0030838) |
| 0.2 |
0.5 |
GO:1900186 |
negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) |
| 0.2 |
0.2 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.2 |
2.3 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.2 |
0.5 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.2 |
3.2 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.2 |
2.4 |
GO:0031297 |
replication fork processing(GO:0031297) |
| 0.2 |
0.5 |
GO:0048597 |
post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.2 |
1.3 |
GO:0031053 |
primary miRNA processing(GO:0031053) |
| 0.2 |
2.3 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.2 |
1.2 |
GO:0071459 |
protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 |
0.7 |
GO:1900016 |
negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.1 |
1.0 |
GO:0045830 |
positive regulation of isotype switching(GO:0045830) |
| 0.1 |
0.4 |
GO:0090370 |
negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
2.1 |
GO:0030261 |
chromosome condensation(GO:0030261) |
| 0.1 |
1.3 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.1 |
0.3 |
GO:0007097 |
nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 |
0.4 |
GO:0042796 |
snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 |
1.7 |
GO:0021924 |
cell proliferation in external granule layer(GO:0021924) cerebellar granule cell precursor proliferation(GO:0021930) |
| 0.1 |
2.5 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.1 |
0.7 |
GO:0045671 |
negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 |
2.8 |
GO:0034198 |
cellular response to amino acid starvation(GO:0034198) |
| 0.1 |
10.6 |
GO:0000082 |
G1/S transition of mitotic cell cycle(GO:0000082) |
| 0.1 |
0.5 |
GO:0006999 |
nuclear pore organization(GO:0006999) |
| 0.1 |
0.1 |
GO:0060355 |
positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 |
1.9 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.1 |
2.6 |
GO:0051084 |
'de novo' protein folding(GO:0006458) 'de novo' posttranslational protein folding(GO:0051084) |
| 0.1 |
12.8 |
GO:0019722 |
calcium-mediated signaling(GO:0019722) |
| 0.1 |
1.7 |
GO:0009048 |
dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 |
4.1 |
GO:0007200 |
phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.1 |
1.5 |
GO:0006353 |
DNA-templated transcription, termination(GO:0006353) |
| 0.1 |
0.2 |
GO:0036166 |
phenotypic switching(GO:0036166) |
| 0.1 |
0.5 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 0.1 |
0.3 |
GO:0032471 |
negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.1 |
0.4 |
GO:0034351 |
negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 |
1.6 |
GO:0048048 |
embryonic eye morphogenesis(GO:0048048) |
| 0.1 |
0.9 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.1 |
1.2 |
GO:0046782 |
regulation of viral transcription(GO:0046782) |
| 0.1 |
2.2 |
GO:0032781 |
positive regulation of ATPase activity(GO:0032781) |
| 0.1 |
2.6 |
GO:0060325 |
face morphogenesis(GO:0060325) |
| 0.1 |
1.2 |
GO:0000394 |
RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
0.9 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
1.3 |
GO:0034453 |
microtubule anchoring(GO:0034453) |
| 0.1 |
1.5 |
GO:0002088 |
lens development in camera-type eye(GO:0002088) |
| 0.1 |
0.3 |
GO:0048665 |
neuron fate specification(GO:0048665) |
| 0.1 |
2.5 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.5 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
1.6 |
GO:0043968 |
histone H2A acetylation(GO:0043968) |
| 0.1 |
1.0 |
GO:0045647 |
negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 |
0.8 |
GO:0030728 |
ovulation(GO:0030728) |
| 0.1 |
1.5 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 |
0.9 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 |
2.3 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 |
4.0 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.1 |
3.2 |
GO:0009168 |
purine nucleoside monophosphate biosynthetic process(GO:0009127) purine ribonucleoside monophosphate biosynthetic process(GO:0009168) |
| 0.1 |
1.0 |
GO:0032479 |
regulation of type I interferon production(GO:0032479) interferon-beta production(GO:0032608) regulation of interferon-beta production(GO:0032648) |
| 0.1 |
1.0 |
GO:0006107 |
oxaloacetate metabolic process(GO:0006107) |
| 0.1 |
1.8 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.1 |
2.9 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 |
0.5 |
GO:0090244 |
Wnt signaling pathway involved in somitogenesis(GO:0090244) |
| 0.1 |
0.5 |
GO:0045721 |
negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 |
3.1 |
GO:0015804 |
neutral amino acid transport(GO:0015804) |
| 0.1 |
1.0 |
GO:0034314 |
Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 |
0.6 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 |
2.7 |
GO:0001885 |
endothelial cell development(GO:0001885) |
| 0.1 |
0.1 |
GO:0046851 |
negative regulation of bone resorption(GO:0045779) negative regulation of bone remodeling(GO:0046851) |
| 0.1 |
1.9 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 |
0.7 |
GO:0099625 |
regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.1 |
0.5 |
GO:0071157 |
negative regulation of cell cycle arrest(GO:0071157) |
| 0.1 |
1.9 |
GO:2001238 |
positive regulation of extrinsic apoptotic signaling pathway(GO:2001238) |
| 0.1 |
0.3 |
GO:0097428 |
protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 |
1.7 |
GO:0043550 |
regulation of lipid kinase activity(GO:0043550) |
| 0.1 |
2.0 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 |
0.3 |
GO:0000290 |
deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 |
0.1 |
GO:0038163 |
thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 |
1.4 |
GO:0001947 |
heart looping(GO:0001947) |
| 0.1 |
0.5 |
GO:0006610 |
ribosomal protein import into nucleus(GO:0006610) |
| 0.1 |
1.0 |
GO:0051898 |
negative regulation of protein kinase B signaling(GO:0051898) |
| 0.1 |
1.4 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.1 |
0.8 |
GO:0050892 |
intestinal absorption(GO:0050892) |
| 0.1 |
0.5 |
GO:0099515 |
actin filament-based transport(GO:0099515) |
| 0.1 |
0.9 |
GO:0043162 |
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 |
1.3 |
GO:0007020 |
microtubule nucleation(GO:0007020) |
| 0.1 |
0.8 |
GO:0048873 |
homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 |
0.7 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.0 |
0.8 |
GO:0060765 |
regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 |
0.5 |
GO:0010867 |
positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 |
7.3 |
GO:0007266 |
Rho protein signal transduction(GO:0007266) |
| 0.0 |
0.3 |
GO:1903069 |
regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 |
0.5 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.8 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.0 |
0.9 |
GO:0030512 |
negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 |
1.3 |
GO:0045494 |
photoreceptor cell maintenance(GO:0045494) |
| 0.0 |
0.1 |
GO:0060971 |
embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 |
1.5 |
GO:0006405 |
RNA export from nucleus(GO:0006405) |
| 0.0 |
1.5 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
1.2 |
GO:0071277 |
cellular response to calcium ion(GO:0071277) |
| 0.0 |
3.2 |
GO:0048864 |
stem cell development(GO:0048864) |
| 0.0 |
0.2 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.1 |
GO:0051533 |
positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 |
0.3 |
GO:0017196 |
N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 |
0.1 |
GO:0048570 |
notochord morphogenesis(GO:0048570) |
| 0.0 |
0.3 |
GO:0034219 |
carbohydrate transmembrane transport(GO:0034219) |
| 0.0 |
0.3 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.4 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.0 |
2.1 |
GO:0048863 |
stem cell differentiation(GO:0048863) |
| 0.0 |
0.6 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.0 |
0.2 |
GO:0015675 |
nickel cation transport(GO:0015675) |
| 0.0 |
0.3 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.0 |
0.2 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
3.5 |
GO:0010466 |
negative regulation of peptidase activity(GO:0010466) |
| 0.0 |
0.3 |
GO:0060351 |
cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 |
1.9 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
1.2 |
GO:0048066 |
developmental pigmentation(GO:0048066) |
| 0.0 |
0.5 |
GO:0030865 |
cortical cytoskeleton organization(GO:0030865) |
| 0.0 |
1.3 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
2.2 |
GO:0045137 |
development of primary sexual characteristics(GO:0045137) |
| 0.0 |
0.5 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
0.7 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.0 |
1.5 |
GO:0030326 |
embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 |
0.8 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.9 |
GO:0000387 |
spliceosomal snRNP assembly(GO:0000387) |
| 0.0 |
0.0 |
GO:0044065 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) regulation of respiratory system process(GO:0044065) |
| 0.0 |
0.5 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.6 |
GO:1900046 |
regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 |
0.3 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.3 |
GO:0035272 |
exocrine system development(GO:0035272) |
| 0.0 |
0.4 |
GO:1903146 |
regulation of mitophagy(GO:1903146) |
| 0.0 |
0.2 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.0 |
0.6 |
GO:0009267 |
cellular response to starvation(GO:0009267) |
| 0.0 |
1.3 |
GO:0007519 |
skeletal muscle tissue development(GO:0007519) |
| 0.0 |
0.1 |
GO:0033353 |
S-adenosylmethionine cycle(GO:0033353) |
| 0.0 |
0.5 |
GO:0032006 |
regulation of TOR signaling(GO:0032006) |
| 0.0 |
0.6 |
GO:0006413 |
translational initiation(GO:0006413) |