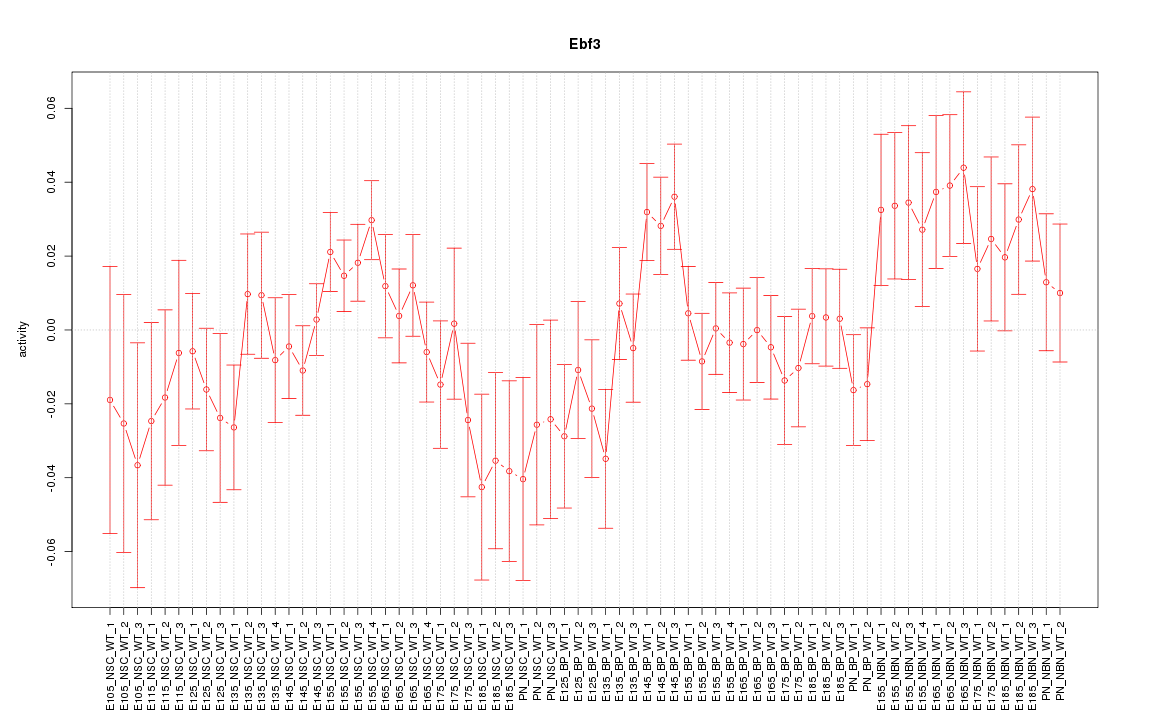
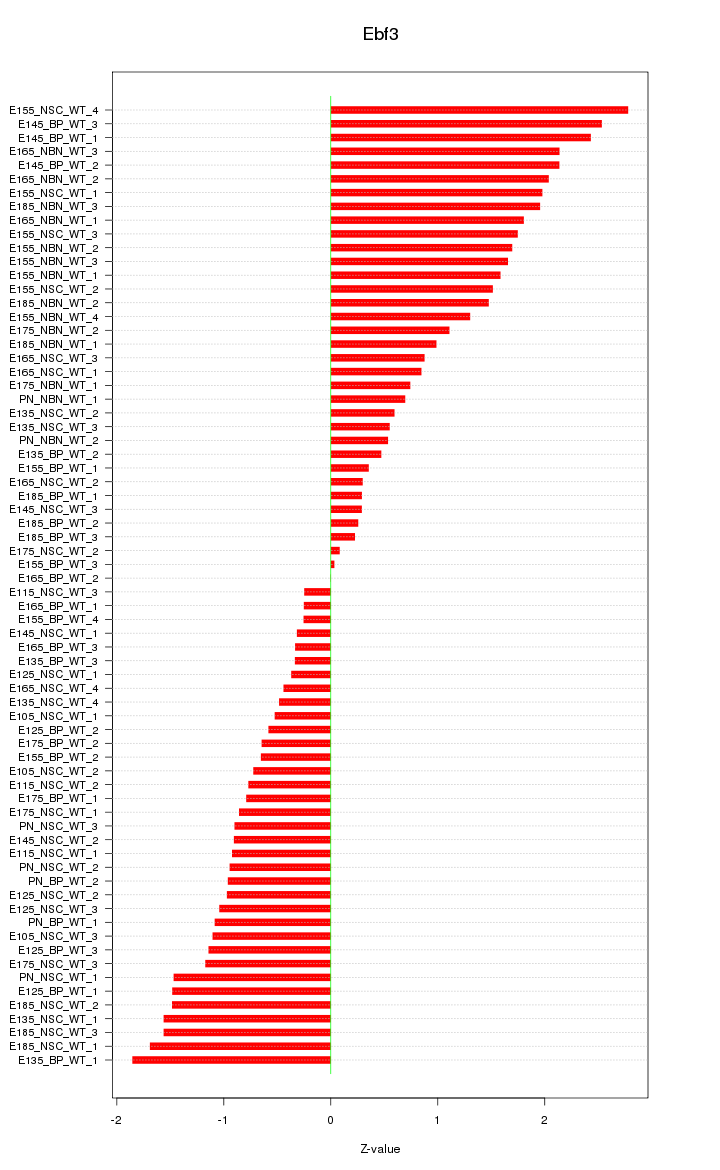
Motif ID: Ebf3
Z-value: 1.209
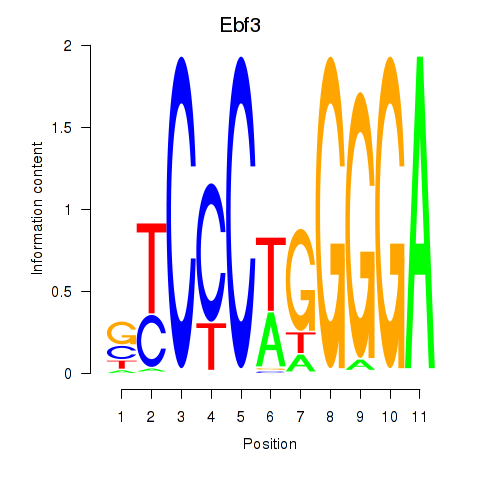
Transcription factors associated with Ebf3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ebf3 | ENSMUSG00000010476.7 | Ebf3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ebf3 | mm10_v2_chr7_-_137314394_137314445 | -0.10 | 3.9e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 4.0 | 16.2 | GO:1901204 | positive regulation of guanylate cyclase activity(GO:0031284) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 4.0 | 12.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 2.6 | 42.2 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 2.3 | 9.3 | GO:0021564 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 1.8 | 28.9 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.7 | 8.4 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 1.6 | 6.5 | GO:1904529 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 1.6 | 12.6 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.4 | 5.5 | GO:2000551 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 1.4 | 4.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 1.3 | 16.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 1.3 | 7.9 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 1.3 | 3.8 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.2 | 7.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.2 | 7.3 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 1.2 | 9.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 1.1 | 3.3 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.9 | 4.7 | GO:0030091 | protein repair(GO:0030091) |
| 0.9 | 2.8 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.9 | 2.6 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.8 | 6.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.8 | 4.6 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 2.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.7 | 5.3 | GO:0097460 | ferrous iron import into cell(GO:0097460) |
| 0.7 | 3.3 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.6 | 2.4 | GO:0042373 | vitamin K metabolic process(GO:0042373) |
| 0.6 | 15.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.6 | 1.7 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.6 | 6.2 | GO:0051386 | regulation of neurotrophin TRK receptor signaling pathway(GO:0051386) |
| 0.5 | 2.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.5 | 2.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 2.1 | GO:1900451 | striatal medium spiny neuron differentiation(GO:0021773) positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.5 | 1.5 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.5 | 4.5 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.5 | 2.4 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.5 | 2.3 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.4 | 1.7 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.4 | 0.8 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.4 | 2.7 | GO:0071732 | cellular response to nitric oxide(GO:0071732) |
| 0.4 | 5.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.4 | 5.3 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.4 | 6.3 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.3 | 1.7 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.3 | 1.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 5.8 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 6.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.3 | 0.3 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.3 | 1.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 1.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 10.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.3 | 6.7 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.3 | 2.6 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.3 | 1.9 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.3 | 1.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.3 | 1.0 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.2 | 4.0 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.2 | 1.2 | GO:0070627 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) ferrous iron import(GO:0070627) |
| 0.2 | 1.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 3.5 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 1.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.2 | 9.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 5.1 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.2 | 3.4 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.2 | 4.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 0.8 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.2 | 0.6 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 1.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.6 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.2 | 3.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 1.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.2 | 4.4 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) |
| 0.2 | 0.8 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.2 | 0.8 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.2 | 1.7 | GO:0043206 | extracellular fibril organization(GO:0043206) |
| 0.2 | 3.0 | GO:0001765 | membrane raft assembly(GO:0001765) |
| 0.2 | 0.5 | GO:2000334 | response to linoleic acid(GO:0070543) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.2 | 1.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 3.6 | GO:0048148 | behavioral response to cocaine(GO:0048148) |
| 0.2 | 1.9 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.2 | 2.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 2.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.9 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.7 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.4 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.1 | 1.3 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.1 | 5.6 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.1 | 0.5 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.1 | 4.1 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 4.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 1.2 | GO:0060732 | positive regulation of inositol phosphate biosynthetic process(GO:0060732) |
| 0.1 | 2.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.1 | 1.6 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.1 | 0.2 | GO:0002865 | negative regulation of acute inflammatory response to antigenic stimulus(GO:0002865) |
| 0.1 | 1.8 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 3.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.9 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.5 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.6 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.1 | 0.7 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 | 3.3 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.1 | 1.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.1 | 1.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 1.3 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.1 | 0.4 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.1 | 2.4 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.1 | 1.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.1 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.1 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.1 | 1.3 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 4.5 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.9 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 4.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.5 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.6 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 2.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.1 | 0.7 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.1 | 0.3 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 1.3 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.0 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.5 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 2.9 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 5.2 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.6 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.8 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.2 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 1.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.7 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.3 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.8 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 1.0 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.7 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 1.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.0 | 0.6 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 1.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.3 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 42.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 2.2 | 6.7 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.9 | 18.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 1.5 | 4.6 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 1.5 | 3.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 1.5 | 16.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.2 | 16.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 1.2 | 18.4 | GO:0031045 | dense core granule(GO:0031045) |
| 1.1 | 3.3 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.9 | 5.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 9.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.8 | 10.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.6 | 5.1 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.6 | 1.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.6 | 2.8 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.5 | 4.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.4 | 17.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.4 | 3.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 2.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 4.0 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 4.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.3 | 1.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.3 | 2.3 | GO:0097433 | dense body(GO:0097433) |
| 0.3 | 6.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 1.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 3.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 7.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 23.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.2 | 6.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 2.3 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 9.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.9 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 13.5 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 5.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 6.0 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.2 | 12.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.3 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 4.5 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.3 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 7.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 1.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 3.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.1 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.9 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 2.3 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 2.1 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 2.1 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.6 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 5.9 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.0 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.7 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 2.4 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 31.5 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.7 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.5 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 1.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.0 | GO:0035869 | ciliary transition zone(GO:0035869) |
| 0.0 | 1.3 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 10.0 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 3.0 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 6.6 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.7 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.8 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 2.3 | GO:0005764 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 0.8 | GO:0005814 | centriole(GO:0005814) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 16.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 3.3 | 3.3 | GO:0086056 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 3.1 | 15.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 1.6 | 36.9 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.5 | 4.5 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 1.5 | 16.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 1.3 | 5.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 1.3 | 6.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.2 | 7.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.1 | 6.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.0 | 4.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 1.0 | 13.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.8 | 6.3 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) |
| 0.7 | 4.4 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.7 | 2.2 | GO:0070905 | serine binding(GO:0070905) |
| 0.7 | 9.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.6 | 1.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.6 | 2.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.5 | 2.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.5 | 6.6 | GO:0031005 | filamin binding(GO:0031005) |
| 0.5 | 3.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.5 | 4.7 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 1.3 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.4 | 3.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.4 | 16.5 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 7.0 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 3.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.4 | 9.6 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.4 | 7.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.3 | 8.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.3 | 2.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.3 | 3.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 2.4 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.3 | 2.3 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.3 | 1.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.3 | 6.0 | GO:0017161 | phosphohistidine phosphatase activity(GO:0008969) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) NADP phosphatase activity(GO:0019178) 5-amino-6-(5-phosphoribitylamino)uracil phosphatase activity(GO:0043726) phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) inositol-1,3,4-trisphosphate 1-phosphatase activity(GO:0052829) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) phosphatidylinositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052867) IDP phosphatase activity(GO:1990003) |
| 0.3 | 1.7 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.3 | 2.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.3 | 2.7 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 5.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.2 | 1.2 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 1.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 8.7 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.2 | 6.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.9 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.2 | 1.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.2 | 1.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.2 | 1.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 3.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 3.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 0.5 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.2 | 3.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 3.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 4.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.7 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 1.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.1 | 1.6 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 4.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 6.0 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 1.7 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 1.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 2.8 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 7.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 4.5 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.1 | 8.3 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.1 | 0.5 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.1 | 1.0 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.1 | 3.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 5.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 1.9 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 3.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 4.5 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 1.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 1.0 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 1.4 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 2.6 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.0 | 2.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.6 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 2.1 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 4.2 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |