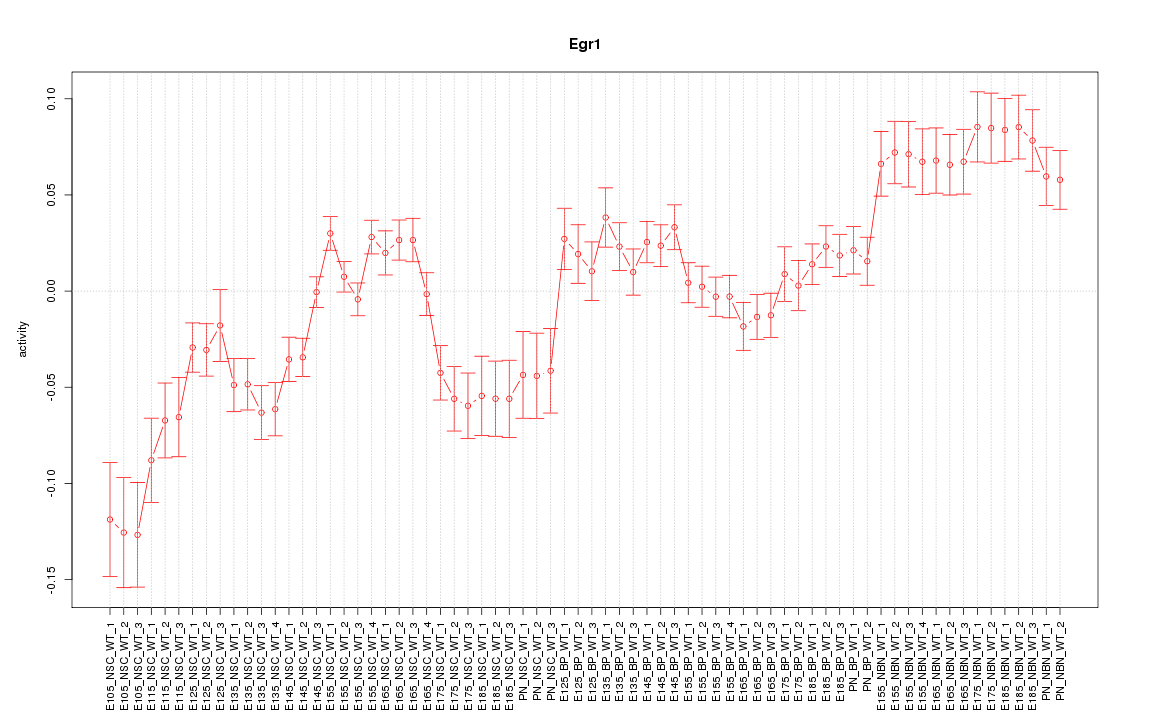
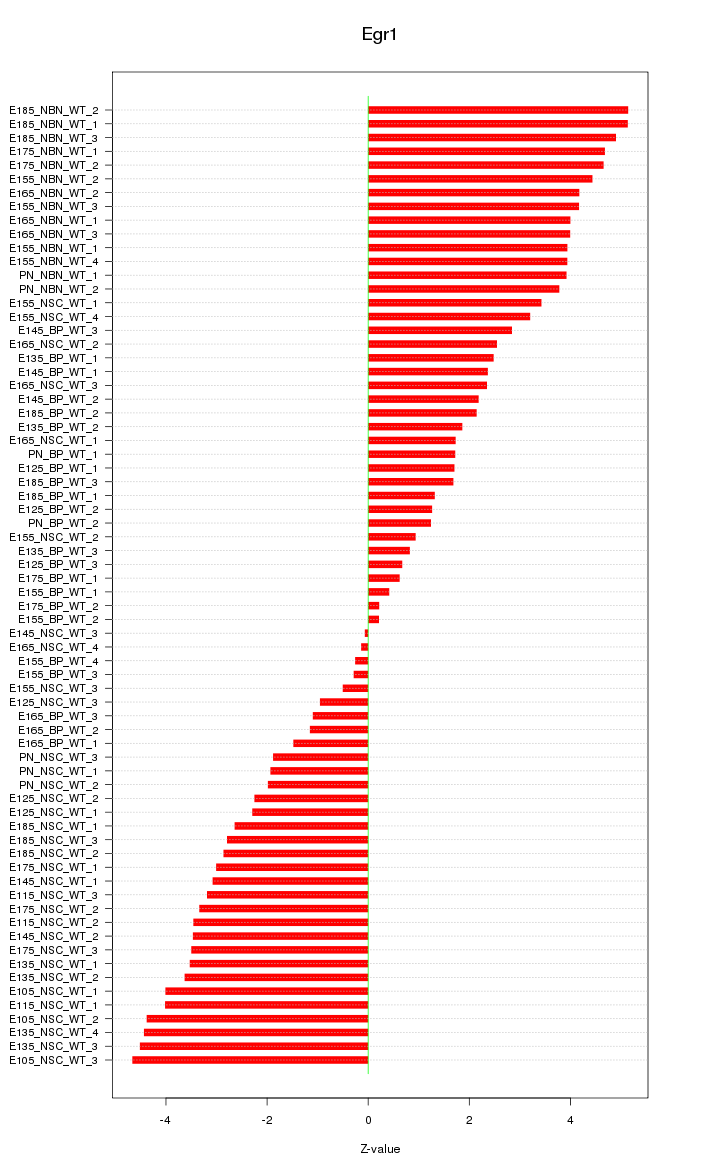
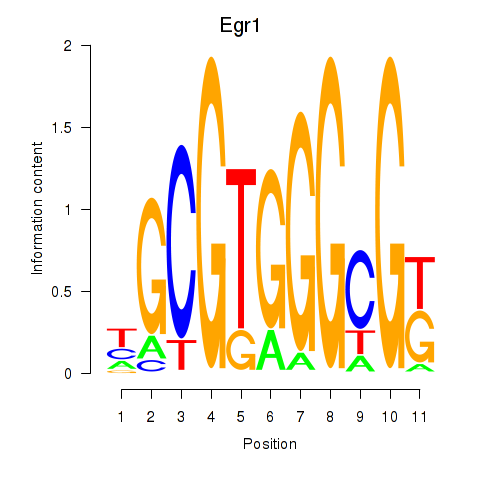
Motif ID: Egr1
Z-value: 2.974
Transcription factors associated with Egr1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Egr1 | ENSMUSG00000038418.7 | Egr1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr1 | mm10_v2_chr18_+_34861200_34861215 | -0.37 | 1.5e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 39.1 | 117.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 15.0 | 45.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 10.5 | 42.0 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 10.2 | 51.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 9.3 | 28.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 9.0 | 27.0 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 7.8 | 46.6 | GO:0032796 | uropod organization(GO:0032796) |
| 7.2 | 36.2 | GO:2000325 | regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000325) positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 7.1 | 21.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 6.8 | 20.5 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 5.2 | 31.1 | GO:1900451 | positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 5.2 | 15.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 5.1 | 25.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 5.1 | 60.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 5.0 | 19.8 | GO:0010040 | response to iron(II) ion(GO:0010040) positive regulation of hydrogen peroxide metabolic process(GO:0010726) negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) cellular response to copper ion(GO:0071280) regulation of peroxidase activity(GO:2000468) |
| 4.9 | 24.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 4.5 | 13.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 4.3 | 21.7 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 4.3 | 12.8 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 4.1 | 16.5 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 3.7 | 18.6 | GO:0090292 | nuclear migration along microfilament(GO:0031022) nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 3.6 | 10.9 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 3.5 | 21.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 3.5 | 10.4 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 3.4 | 10.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 3.3 | 16.3 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 3.1 | 9.4 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 3.0 | 14.9 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 2.8 | 22.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 2.8 | 41.6 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 2.7 | 18.8 | GO:1903423 | positive regulation of synaptic vesicle recycling(GO:1903423) |
| 2.6 | 15.6 | GO:1901524 | regulation of autophagosome maturation(GO:1901096) regulation of macromitophagy(GO:1901524) negative regulation of macromitophagy(GO:1901525) |
| 2.6 | 18.0 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 2.5 | 12.7 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 2.5 | 15.0 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 2.5 | 32.5 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 2.5 | 12.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 2.5 | 14.7 | GO:0019695 | choline metabolic process(GO:0019695) |
| 2.4 | 7.2 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) protein poly-ADP-ribosylation(GO:0070212) |
| 2.4 | 7.2 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 2.4 | 9.5 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 2.3 | 18.6 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 2.3 | 23.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 2.2 | 6.7 | GO:1990314 | pyrimidine-containing compound transmembrane transport(GO:0072531) cellular response to insulin-like growth factor stimulus(GO:1990314) |
| 2.2 | 8.7 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 2.1 | 10.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 2.1 | 8.6 | GO:0070305 | response to cGMP(GO:0070305) cellular response to cGMP(GO:0071321) |
| 2.1 | 4.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 2.1 | 6.3 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 2.0 | 16.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 2.0 | 6.1 | GO:0030070 | insulin processing(GO:0030070) |
| 1.9 | 11.4 | GO:0097491 | trigeminal nerve morphogenesis(GO:0021636) trigeminal nerve structural organization(GO:0021637) trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) neural crest cell migration involved in autonomic nervous system development(GO:1901166) |
| 1.9 | 20.8 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 1.9 | 9.4 | GO:2001286 | regulation of caveolin-mediated endocytosis(GO:2001286) |
| 1.8 | 11.0 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 1.8 | 9.1 | GO:0097343 | positive regulation of T cell receptor signaling pathway(GO:0050862) ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 1.8 | 25.4 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.7 | 6.7 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 1.6 | 28.8 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 1.6 | 23.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 1.6 | 15.7 | GO:0043084 | penile erection(GO:0043084) |
| 1.5 | 4.6 | GO:0071336 | positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 1.5 | 7.3 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 1.4 | 5.5 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 1.4 | 4.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.3 | 14.8 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.3 | 13.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 1.3 | 7.8 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 1.3 | 11.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 1.3 | 5.2 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 1.3 | 3.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 1.2 | 3.6 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 1.2 | 4.8 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 1.2 | 15.4 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 1.2 | 8.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 1.2 | 3.5 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 1.2 | 6.9 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 1.1 | 6.7 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 1.1 | 10.1 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 1.1 | 3.4 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 1.1 | 3.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) phosphate ion transmembrane transport(GO:0035435) |
| 1.1 | 12.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 1.1 | 4.5 | GO:0032026 | response to magnesium ion(GO:0032026) |
| 1.1 | 7.7 | GO:2000786 | positive regulation of vacuole organization(GO:0044090) positive regulation of autophagosome assembly(GO:2000786) |
| 1.0 | 7.1 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 1.0 | 1.0 | GO:1903061 | regulation of protein lipidation(GO:1903059) positive regulation of protein lipidation(GO:1903061) |
| 1.0 | 28.0 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.9 | 8.5 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.9 | 5.6 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.9 | 10.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.9 | 23.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.9 | 9.0 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.9 | 12.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.9 | 2.6 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.9 | 5.1 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.9 | 12.8 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.8 | 3.4 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.8 | 5.7 | GO:0015862 | uridine transport(GO:0015862) |
| 0.8 | 11.3 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.8 | 4.0 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.8 | 6.4 | GO:0014898 | muscle hypertrophy in response to stress(GO:0003299) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.8 | 6.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.8 | 6.1 | GO:2001224 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) positive regulation of neuron migration(GO:2001224) |
| 0.7 | 5.2 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.7 | 5.9 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.7 | 5.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.7 | 2.2 | GO:0071873 | response to norepinephrine(GO:0071873) |
| 0.7 | 12.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.7 | 18.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.7 | 2.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.7 | 26.4 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.7 | 4.1 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.7 | 1.4 | GO:0060527 | prostate glandular acinus morphogenesis(GO:0060526) prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis(GO:0060527) |
| 0.7 | 4.8 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.7 | 5.5 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.7 | 6.8 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.7 | 17.2 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.6 | 7.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.6 | 12.4 | GO:0061157 | mRNA destabilization(GO:0061157) |
| 0.6 | 9.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.6 | 4.9 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.6 | 10.8 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.6 | 4.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.6 | 22.3 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.6 | 3.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 3.9 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.6 | 4.5 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.6 | 10.5 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.5 | 6.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.5 | 2.7 | GO:2000807 | regulation of synaptic vesicle clustering(GO:2000807) |
| 0.5 | 3.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.5 | 2.1 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.5 | 7.8 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.5 | 2.6 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.5 | 19.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.5 | 3.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.5 | 38.6 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.4 | 3.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.4 | 3.0 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.4 | 3.8 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.4 | 1.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.4 | 7.5 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.4 | 8.4 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.4 | 9.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.4 | 9.3 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.4 | 13.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.4 | 10.6 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.4 | 4.7 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.4 | 1.4 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.4 | 1.1 | GO:0006106 | fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.4 | 8.2 | GO:0042755 | eating behavior(GO:0042755) |
| 0.4 | 1.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.3 | 1.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.3 | 2.4 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.3 | 8.5 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.3 | 1.7 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 1.7 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) negative regulation of filopodium assembly(GO:0051490) |
| 0.3 | 10.0 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.3 | 14.6 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.3 | 3.7 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.3 | 2.6 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.3 | 9.9 | GO:1900078 | positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.3 | 1.7 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 2.9 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.3 | 8.4 | GO:2001240 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.3 | 1.6 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.3 | 8.9 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.3 | 0.9 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.3 | 5.3 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 7.0 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.3 | 22.0 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.3 | 4.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.3 | 10.7 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.3 | 8.5 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.3 | 23.6 | GO:0099643 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.3 | 16.6 | GO:2001237 | negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.3 | 1.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.3 | 17.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.3 | 34.4 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.3 | 1.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.3 | 3.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.3 | 6.1 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.3 | 0.8 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.3 | 0.8 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 2.0 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.2 | 1.5 | GO:0030578 | PML body organization(GO:0030578) |
| 0.2 | 27.2 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.2 | 4.4 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.2 | 2.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.2 | 22.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.2 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 4.5 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.2 | 1.0 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 | 9.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.2 | 5.6 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 2.1 | GO:1990173 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.2 | 4.7 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.2 | 2.7 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 8.5 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.2 | 18.3 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.1 | 4.2 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 18.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.1 | 0.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 3.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 1.7 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 0.9 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 2.8 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.1 | 1.8 | GO:1901798 | positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.1 | 5.3 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.9 | GO:0009988 | cell-cell recognition(GO:0009988) |
| 0.1 | 1.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.1 | 2.5 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.1 | 2.1 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 3.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 3.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.1 | 0.4 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 15.3 | GO:0006457 | protein folding(GO:0006457) |
| 0.1 | 1.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 1.4 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 0.1 | 3.1 | GO:0042770 | signal transduction in response to DNA damage(GO:0042770) |
| 0.1 | 0.4 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.6 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 1.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 2.3 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 1.4 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 9.9 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.1 | 0.7 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.8 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 3.2 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.0 | 2.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 4.2 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.0 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 | 1.6 | GO:0046939 | nucleoside diphosphate phosphorylation(GO:0006165) nucleotide phosphorylation(GO:0046939) |
| 0.0 | 1.9 | GO:0016197 | endosomal transport(GO:0016197) |
| 0.0 | 0.4 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.5 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 24.6 | GO:0090537 | CERF complex(GO:0090537) |
| 5.2 | 31.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 5.1 | 20.5 | GO:0044307 | dendritic branch(GO:0044307) |
| 5.0 | 65.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 5.0 | 19.8 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 3.9 | 23.2 | GO:0008091 | spectrin(GO:0008091) |
| 3.8 | 11.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 3.7 | 118.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 3.4 | 27.0 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 3.2 | 19.5 | GO:1990635 | proximal dendrite(GO:1990635) |
| 2.4 | 9.4 | GO:0043511 | inhibin complex(GO:0043511) |
| 2.2 | 42.0 | GO:0002102 | podosome(GO:0002102) |
| 2.1 | 18.6 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 1.9 | 46.7 | GO:0001891 | phagocytic cup(GO:0001891) |
| 1.8 | 7.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 1.7 | 15.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 1.6 | 23.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 1.6 | 4.7 | GO:0044194 | cytolytic granule(GO:0044194) |
| 1.6 | 31.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 1.5 | 131.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 1.5 | 9.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 1.3 | 16.3 | GO:0034361 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 1.2 | 45.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 1.2 | 62.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 1.2 | 4.7 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 1.2 | 3.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 1.1 | 6.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 1.1 | 13.5 | GO:0031105 | septin complex(GO:0031105) |
| 1.1 | 10.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 1.1 | 5.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.9 | 6.3 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.9 | 5.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.9 | 28.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.8 | 13.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.8 | 4.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.8 | 6.3 | GO:0005902 | microvillus(GO:0005902) |
| 0.8 | 3.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.8 | 3.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.7 | 9.1 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.7 | 8.0 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.6 | 4.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.6 | 5.0 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.6 | 150.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.6 | 8.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.6 | 9.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.6 | 2.8 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.6 | 7.8 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.5 | 3.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.5 | 70.2 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.5 | 2.7 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 3.2 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.5 | 5.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.5 | 9.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.4 | 10.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.4 | 6.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.4 | 30.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.4 | 11.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.4 | 6.9 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.3 | 4.5 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.3 | 29.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.3 | 8.0 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.3 | 3.4 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 6.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.3 | 10.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.3 | 14.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.3 | 46.8 | GO:0005769 | early endosome(GO:0005769) |
| 0.3 | 119.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.3 | 1.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 2.4 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.3 | 31.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.2 | 10.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 8.2 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.2 | 17.0 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.2 | 5.9 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.2 | 50.4 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.2 | 3.0 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.2 | 2.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.2 | 3.5 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 2.0 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.2 | 1.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 4.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 2.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 19.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.1 | 3.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.1 | 4.5 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 5.5 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 2.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.1 | 1.5 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 8.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 3.9 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 7.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 6.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 1.2 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 7.1 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.1 | 9.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 2.8 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.1 | 0.6 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 2.1 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.2 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 9.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.1 | 1.7 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.1 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 11.1 | GO:0016604 | nuclear body(GO:0016604) |
| 0.1 | 26.5 | GO:0005773 | vacuole(GO:0005773) |
| 0.1 | 7.5 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.1 | 1.4 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 7.7 | GO:0031253 | cell projection membrane(GO:0031253) |
| 0.1 | 4.7 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 25.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 0.7 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.7 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 7.1 | GO:0005929 | cilium(GO:0005929) |
| 0.0 | 0.7 | GO:0030426 | growth cone(GO:0030426) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.0 | 45.0 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 8.2 | 24.6 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 5.7 | 137.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 5.6 | 61.8 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 5.4 | 16.3 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) glycoprotein transporter activity(GO:0034437) |
| 5.2 | 42.0 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 4.7 | 46.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 4.5 | 18.2 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 3.9 | 23.4 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 3.7 | 37.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 3.7 | 14.7 | GO:0004104 | cholinesterase activity(GO:0004104) choline binding(GO:0033265) |
| 3.7 | 51.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 3.7 | 18.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 3.4 | 13.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 3.2 | 12.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 3.0 | 14.9 | GO:0004359 | glutaminase activity(GO:0004359) |
| 2.8 | 8.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 2.7 | 8.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 2.6 | 15.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.6 | 72.4 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 2.5 | 10.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 2.4 | 14.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 2.3 | 11.3 | GO:0018121 | imidazoleglycerol-phosphate synthase activity(GO:0000107) NAD(P)-cysteine ADP-ribosyltransferase activity(GO:0018071) NAD(P)-asparagine ADP-ribosyltransferase activity(GO:0018121) NAD(P)-serine ADP-ribosyltransferase activity(GO:0018127) 7-cyano-7-deazaguanine tRNA-ribosyltransferase activity(GO:0043867) purine deoxyribosyltransferase activity(GO:0044102) |
| 2.0 | 7.8 | GO:2001069 | glycogen binding(GO:2001069) |
| 2.0 | 23.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 2.0 | 5.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 1.9 | 9.3 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 1.9 | 20.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.8 | 9.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.8 | 16.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 1.8 | 5.3 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 1.8 | 5.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 1.7 | 8.6 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 1.7 | 13.6 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 1.7 | 9.9 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 1.6 | 4.9 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 1.6 | 23.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 1.5 | 31.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 1.4 | 6.8 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.3 | 6.7 | GO:0097001 | ceramide binding(GO:0097001) |
| 1.3 | 30.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 1.2 | 4.7 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 1.2 | 11.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 1.2 | 4.6 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.1 | 14.8 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.1 | 13.4 | GO:0016868 | intramolecular transferase activity, phosphotransferases(GO:0016868) |
| 1.1 | 25.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 1.1 | 18.0 | GO:0050253 | prenylcysteine methylesterase activity(GO:0010296) 1-oxa-2-oxocycloheptane lactonase activity(GO:0018731) sulfolactone hydrolase activity(GO:0018732) butyrolactone hydrolase activity(GO:0018734) endosulfan lactone lactonase activity(GO:0034892) L-ascorbate 6-phosphate lactonase activity(GO:0035460) Ser-tRNA(Thr) hydrolase activity(GO:0043905) Ala-tRNA(Pro) hydrolase activity(GO:0043906) Cys-tRNA(Pro) hydrolase activity(GO:0043907) Ser(Gly)-tRNA(Ala) hydrolase activity(GO:0043908) all-trans-retinyl-palmitate hydrolase, all-trans-retinol forming activity(GO:0047376) retinyl-palmitate esterase activity(GO:0050253) mannosyl-oligosaccharide 1,6-alpha-mannosidase activity(GO:0052767) mannosyl-oligosaccharide 1,3-alpha-mannosidase activity(GO:0052768) methyl indole-3-acetate esterase activity(GO:0080030) methyl salicylate esterase activity(GO:0080031) methyl jasmonate esterase activity(GO:0080032) |
| 1.0 | 9.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.0 | 11.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 1.0 | 22.6 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 1.0 | 11.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 1.0 | 4.9 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.0 | 15.5 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 1.0 | 2.9 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.9 | 5.4 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.9 | 5.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.9 | 6.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.8 | 37.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.8 | 5.8 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.8 | 6.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.8 | 6.5 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.8 | 18.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.8 | 23.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.8 | 7.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.8 | 3.1 | GO:0005168 | neurotrophin TRK receptor binding(GO:0005167) neurotrophin TRKA receptor binding(GO:0005168) |
| 0.8 | 11.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.7 | 15.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.7 | 4.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.6 | 4.5 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.6 | 7.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.6 | 6.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.6 | 8.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.6 | 3.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.6 | 5.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.6 | 65.0 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.5 | 2.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.5 | 12.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.5 | 12.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.5 | 10.1 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.5 | 3.0 | GO:0036122 | BMP binding(GO:0036122) |
| 0.5 | 12.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.5 | 12.7 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.5 | 33.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.5 | 10.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.5 | 1.8 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 2.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.4 | 5.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.4 | 14.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.4 | 40.1 | GO:0101005 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.4 | 27.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.4 | 1.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.4 | 21.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 7.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.4 | 3.6 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.4 | 11.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.4 | 1.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.3 | 2.0 | GO:0070728 | leucine binding(GO:0070728) |
| 0.3 | 8.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 3.1 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.3 | 23.2 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.3 | 8.5 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.3 | 1.2 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.3 | 9.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 14.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.3 | 3.5 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.3 | 10.2 | GO:0008748 | N-ethylmaleimide reductase activity(GO:0008748) reduced coenzyme F420 dehydrogenase activity(GO:0043738) sulfur oxygenase reductase activity(GO:0043826) malolactic enzyme activity(GO:0043883) epoxyqueuosine reductase activity(GO:0052693) |
| 0.3 | 1.8 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 7.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 24.7 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.2 | 3.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.2 | 46.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.2 | 4.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 8.4 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 10.9 | GO:0035496 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) O antigen polymerase activity(GO:0008755) lipopolysaccharide-1,6-galactosyltransferase activity(GO:0008921) cellulose synthase activity(GO:0016759) 9-phenanthrol UDP-glucuronosyltransferase activity(GO:0018715) 1-phenanthrol glycosyltransferase activity(GO:0018716) 9-phenanthrol glycosyltransferase activity(GO:0018717) 1,2-dihydroxy-phenanthrene glycosyltransferase activity(GO:0018718) phenanthrol glycosyltransferase activity(GO:0019112) alpha-1,2-galactosyltransferase activity(GO:0031278) dolichyl pyrophosphate Man7GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0033556) endogalactosaminidase activity(GO:0033931) lipopolysaccharide-1,5-galactosyltransferase activity(GO:0035496) dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase activity(GO:0042283) inositol phosphoceramide synthase activity(GO:0045140) alpha-(1->3)-fucosyltransferase activity(GO:0046920) indole-3-butyrate beta-glucosyltransferase activity(GO:0052638) salicylic acid glucosyltransferase (ester-forming) activity(GO:0052639) salicylic acid glucosyltransferase (glucoside-forming) activity(GO:0052640) benzoic acid glucosyltransferase activity(GO:0052641) chondroitin hydrolase activity(GO:0052757) dolichyl-pyrophosphate Man7GlcNAc2 alpha-1,6-mannosyltransferase activity(GO:0052824) cytokinin 9-beta-glucosyltransferase activity(GO:0080062) |
| 0.2 | 4.7 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.2 | 21.9 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.2 | 4.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 0.5 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) |
| 0.2 | 2.2 | GO:0044653 | trehalase activity(GO:0015927) dextrin alpha-glucosidase activity(GO:0044653) starch alpha-glucosidase activity(GO:0044654) beta-glucanase activity(GO:0052736) beta-6-sulfate-N-acetylglucosaminidase activity(GO:0052769) glucan endo-1,4-beta-glucosidase activity(GO:0052859) |
| 0.2 | 6.1 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.2 | 2.0 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 4.6 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.2 | 5.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 14.6 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 1.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 4.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.1 | 13.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 39.0 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 4.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.4 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.9 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.1 | 13.2 | GO:0061659 | ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.1 | 4.2 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 20.4 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 1.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 1.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.7 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 3.5 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 5.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 2.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 4.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 1.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 6.8 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.1 | 5.6 | GO:0019199 | transmembrane receptor protein kinase activity(GO:0019199) |
| 0.1 | 5.8 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 3.2 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.1 | 4.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 2.3 | GO:0001221 | transcription cofactor binding(GO:0001221) |
| 0.1 | 0.8 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 1.0 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 1.5 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 7.5 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 2.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 1.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 4.5 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 0.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |