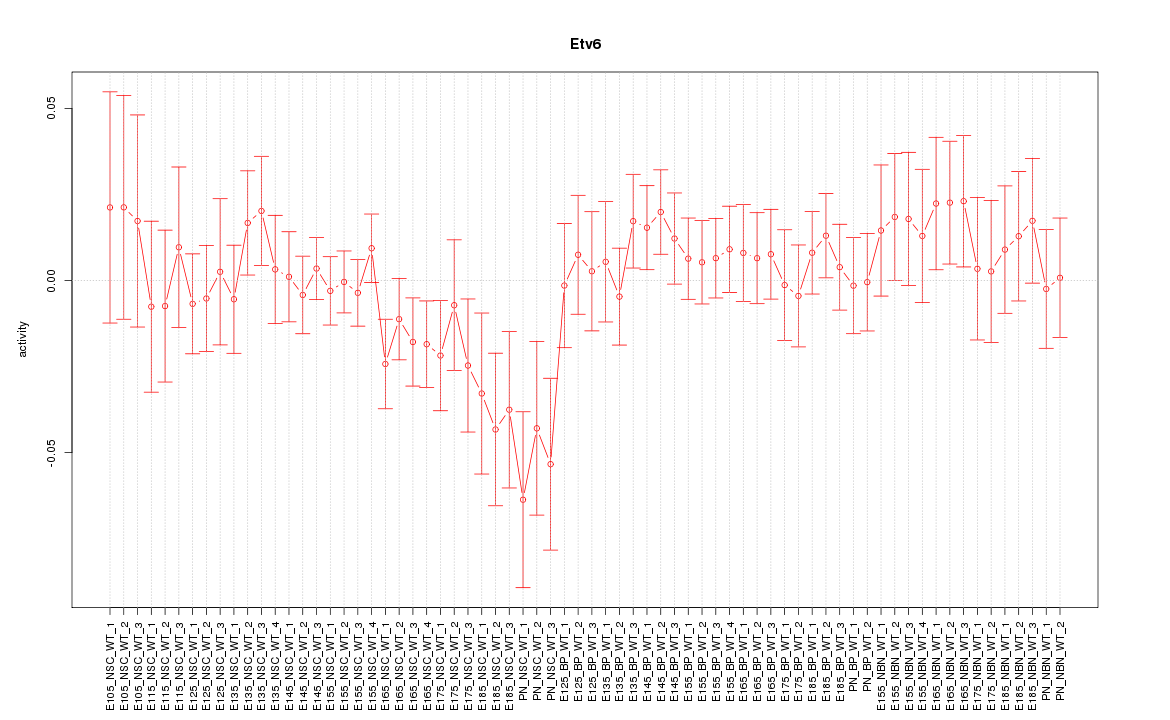
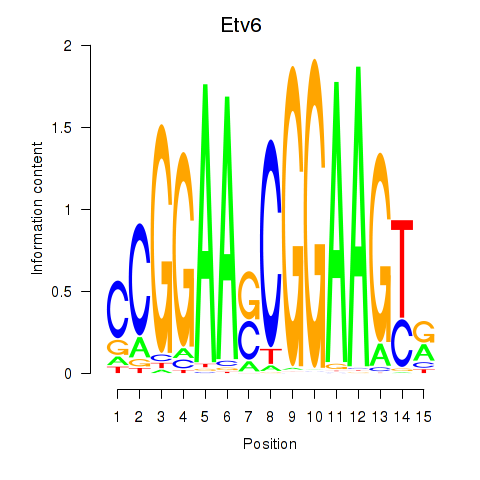
Motif ID: Etv6
Z-value: 0.931
Transcription factors associated with Etv6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Etv6 | ENSMUSG00000030199.10 | Etv6 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Etv6 | mm10_v2_chr6_+_134035691_134035717 | -0.21 | 7.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0060084 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) synaptic transmission involved in micturition(GO:0060084) |
| 0.9 | 2.7 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.8 | 3.2 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) positive regulation of MHC class I biosynthetic process(GO:0045345) |
| 0.8 | 2.4 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.8 | 6.2 | GO:0031053 | production of siRNA involved in RNA interference(GO:0030422) primary miRNA processing(GO:0031053) |
| 0.7 | 2.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.7 | 2.1 | GO:0070944 | neutrophil mediated killing of bacterium(GO:0070944) |
| 0.7 | 6.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.5 | 3.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.5 | 1.5 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.5 | 3.7 | GO:0036506 | maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.5 | 1.8 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.4 | 1.8 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.4 | 1.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.4 | 1.9 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 0.4 | 1.1 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 0.4 | 1.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.3 | 0.7 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.3 | 4.0 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.0 | GO:0046881 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.3 | 1.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.3 | 2.9 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.3 | 0.8 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.3 | 0.8 | GO:0090202 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.7 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.2 | 1.4 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 0.7 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.2 | 0.9 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.2 | 1.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.2 | 2.5 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.2 | 0.7 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 1.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.2 | 1.9 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.2 | 0.6 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 1.0 | GO:1903026 | negative regulation of CREB transcription factor activity(GO:0032792) negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.2 | 1.3 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.2 | 0.6 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.2 | 0.9 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.2 | 2.8 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.2 | 1.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 1.6 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.2 | 1.6 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.2 | 0.9 | GO:0097343 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.2 | 0.8 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.0 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.2 | 5.5 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 1.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 1.1 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.1 | 1.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.6 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.9 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 2.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 2.3 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.1 | 4.9 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 2.1 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 2.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 0.4 | GO:0010920 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) |
| 0.1 | 0.2 | GO:2000152 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) regulation of ubiquitin-specific protease activity(GO:2000152) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 4.2 | GO:0072583 | clathrin-mediated endocytosis(GO:0072583) |
| 0.1 | 1.0 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.1 | 4.4 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.1 | 1.4 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.1 | 1.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 1.3 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.1 | 1.8 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.1 | 0.7 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) intracellular protein transmembrane import(GO:0044743) |
| 0.1 | 0.3 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 1.9 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 1.0 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.7 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 1.8 | GO:1903427 | negative regulation of reactive oxygen species biosynthetic process(GO:1903427) |
| 0.1 | 1.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.6 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 | 0.2 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.4 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 1.0 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.1 | 0.7 | GO:2001197 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 0.5 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) paranodal junction assembly(GO:0030913) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 1.3 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.1 | 0.8 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.4 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.7 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.1 | 1.2 | GO:0031050 | dsRNA fragmentation(GO:0031050) production of miRNAs involved in gene silencing by miRNA(GO:0035196) production of small RNA involved in gene silencing by RNA(GO:0070918) |
| 0.1 | 1.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.1 | 2.1 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 | 1.0 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 2.6 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 0.7 | GO:0014883 | transition between fast and slow fiber(GO:0014883) |
| 0.1 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 1.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.9 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.8 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.1 | 1.4 | GO:0045026 | plasma membrane fusion(GO:0045026) |
| 0.1 | 0.5 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 1.7 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 2.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 3.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.6 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 2.5 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.9 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 1.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.6 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.0 | 0.7 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 1.5 | GO:0050714 | positive regulation of protein secretion(GO:0050714) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 1.0 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.9 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 1.3 | GO:0050954 | sensory perception of sound(GO:0007605) sensory perception of mechanical stimulus(GO:0050954) |
| 0.0 | 1.1 | GO:0043039 | tRNA aminoacylation for protein translation(GO:0006418) tRNA aminoacylation(GO:0043039) |
| 0.0 | 0.1 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.9 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 2.4 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.3 | GO:0030836 | positive regulation of actin filament depolymerization(GO:0030836) |
| 0.0 | 2.2 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.5 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 1.0 | GO:0010762 | regulation of fibroblast migration(GO:0010762) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.5 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.7 | GO:0008286 | insulin receptor signaling pathway(GO:0008286) |
| 0.0 | 0.9 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.0 | 0.4 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 1.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 2.1 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
| 0.0 | 0.7 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.4 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.8 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) |
| 0.0 | 0.5 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 0.6 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 0.0 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.5 | GO:2000816 | negative regulation of mitotic metaphase/anaphase transition(GO:0045841) mitotic spindle checkpoint(GO:0071174) negative regulation of metaphase/anaphase transition of cell cycle(GO:1902100) negative regulation of mitotic sister chromatid separation(GO:2000816) |
| 0.0 | 0.8 | GO:0097190 | apoptotic signaling pathway(GO:0097190) |
| 0.0 | 2.1 | GO:0000398 | RNA splicing, via transesterification reactions(GO:0000375) RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 3.8 | GO:0006412 | translation(GO:0006412) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.2 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.7 | 2.8 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.5 | 1.6 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.5 | 3.7 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.5 | 1.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.5 | 2.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.5 | 4.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.5 | 1.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.4 | 4.2 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.4 | 1.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.3 | 1.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.3 | 1.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.3 | 4.9 | GO:0031045 | dense core granule(GO:0031045) |
| 0.3 | 2.9 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 2.4 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.3 | 2.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.3 | 1.3 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.2 | 0.7 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.2 | 1.5 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 0.7 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.2 | 1.1 | GO:0044439 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) microbody part(GO:0044438) peroxisomal part(GO:0044439) |
| 0.2 | 1.5 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.2 | 1.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.5 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.2 | 1.5 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 0.9 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.2 | 1.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.2 | 4.3 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.2 | 2.4 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.2 | 3.0 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.2 | 1.0 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.2 | 1.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 2.1 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 4.3 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 1.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.3 | GO:0044754 | amphisome(GO:0044753) autolysosome(GO:0044754) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 1.0 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.1 | 2.1 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 1.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.4 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 1.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 1.3 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.1 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.1 | 1.0 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.1 | 0.8 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.1 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.1 | 0.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 4.0 | GO:0005746 | mitochondrial respiratory chain(GO:0005746) |
| 0.1 | 1.0 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 3.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.1 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 3.4 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 3.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.4 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.3 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.4 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 1.3 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.3 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.3 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.5 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 2.1 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.9 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.7 | GO:0044452 | nucleolar part(GO:0044452) |
| 0.0 | 0.5 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 0.6 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.7 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 7.8 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.5 | 1.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.5 | 4.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.4 | 1.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) arginine binding(GO:0034618) |
| 0.4 | 6.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.4 | 1.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.4 | 1.1 | GO:0034952 | 3-octaprenyl-4-hydroxybenzoate carboxy-lyase activity(GO:0008694) 2-hydroxy-3-carboxy-6-oxo-7-methylocta-2,4-dienoate decarboxylase activity(GO:0018791) bis(4-chlorophenyl)acetate decarboxylase activity(GO:0018792) 3,5-dibromo-4-hydroxybenzoate decarboxylase activity(GO:0018793) 2-hydroxyisobutyrate decarboxylase activity(GO:0018794) 2-hydroxy-2-methyl-1,3-dicarbonate decarboxylase activity(GO:0018795) 2-hydroxyisophthalate decarboxylase activity(GO:0034524) dimethylmalonate decarboxylase activity(GO:0034782) 2,4,4-trimethyl-3-oxopentanoate decarboxylase activity(GO:0034853) 4,4-dimethyl-3-oxopentanoate decarboxylase activity(GO:0034854) 2,3,6-trihydroxyisonicotinate decarboxylase activity(GO:0034879) phenanthrene-4,5-dicarboxylate decarboxylase activity(GO:0034923) pyrrole-2-carboxylate decarboxylase activity(GO:0034941) terephthalate decarboxylase activity(GO:0034947) malonate semialdehyde decarboxylase activity(GO:0034952) 5-amino-4-imidazole carboxylate lyase activity(GO:0043727) 2-keto-4-methylthiobutyrate aminotransferase activity(GO:0043728) 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase activity(GO:0051997) |
| 0.3 | 1.0 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.3 | 2.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.3 | 2.9 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.3 | 2.8 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.2 | 0.9 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.2 | 1.5 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 1.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 1.0 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.2 | 2.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.2 | 3.7 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.2 | 1.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.2 | 0.7 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.2 | 0.7 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 0.8 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.2 | 0.6 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 1.0 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.1 | 1.9 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.7 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.8 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.1 | 0.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 1.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 4.9 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 6.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 3.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 3.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 1.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.1 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.9 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 3.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 1.6 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.1 | 1.7 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.7 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.1 | 0.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.3 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 5.1 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA methyltransferase activity(GO:0008649) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) rRNA (uridine) methyltransferase activity(GO:0016436) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.1 | 0.5 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.7 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 1.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.9 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 1.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.6 | GO:0016751 | S-succinyltransferase activity(GO:0016751) |
| 0.1 | 2.1 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 1.0 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 3.8 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.2 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 1.6 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 9.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.9 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.3 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.0 | 0.6 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 3.0 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 1.8 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 1.0 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 1.1 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.5 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 3.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 6.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.3 | GO:0016303 | 1-phosphatidylinositol-3-kinase activity(GO:0016303) |
| 0.0 | 1.6 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.4 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 1.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.6 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 3.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.2 | GO:0034951 | pivalyl-CoA mutase activity(GO:0034784) o-hydroxylaminobenzoate mutase activity(GO:0034951) lupeol synthase activity(GO:0042299) beta-amyrin synthase activity(GO:0042300) baruol synthase activity(GO:0080011) |
| 0.0 | 1.1 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |