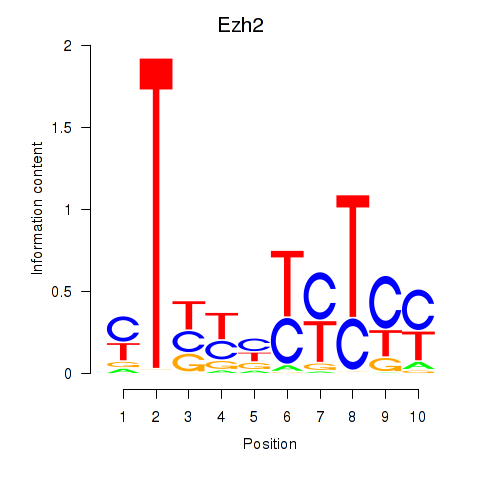
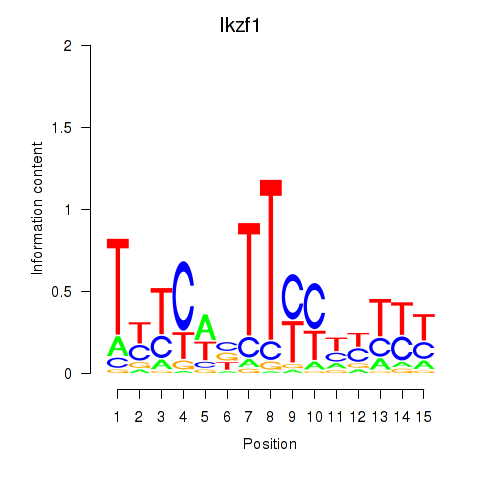
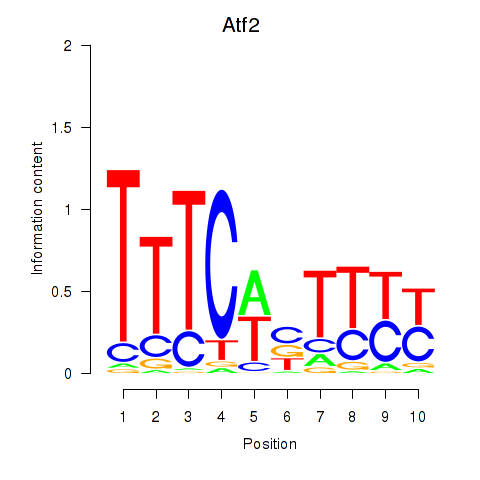
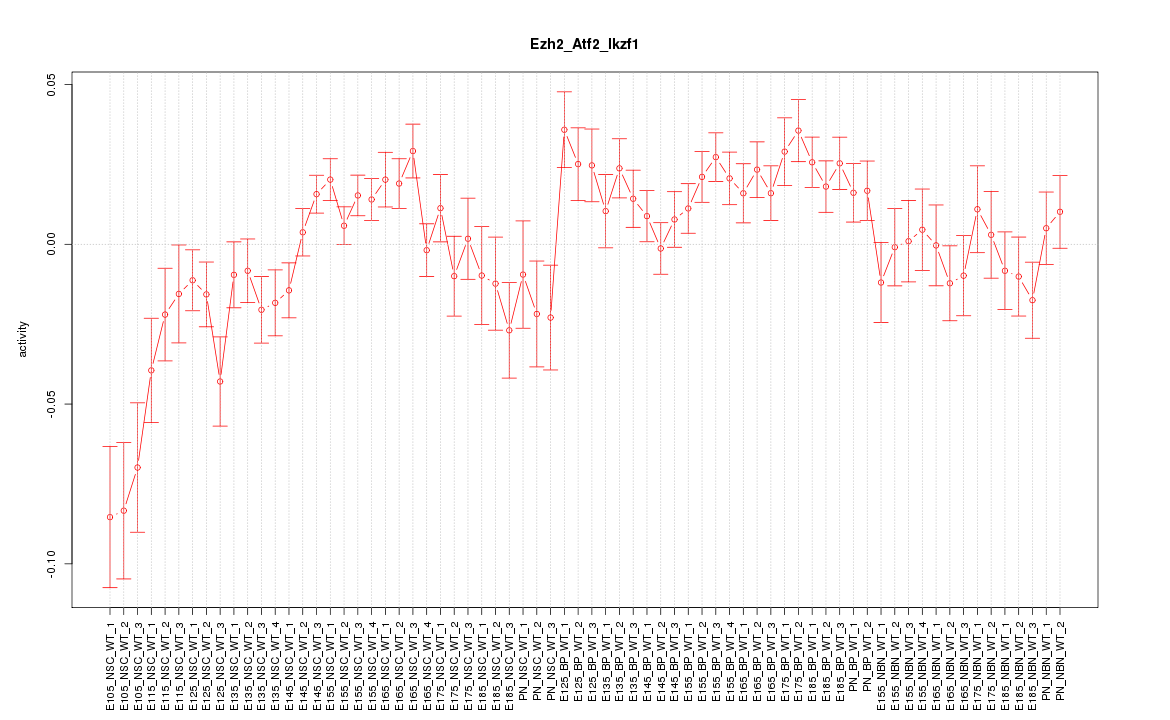
| 16.0 |
64.1 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 4.9 |
24.5 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 4.6 |
23.0 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 4.4 |
13.1 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 3.9 |
11.8 |
GO:0035021 |
negative regulation of Rac protein signal transduction(GO:0035021) |
| 3.6 |
17.8 |
GO:2000481 |
positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 3.4 |
16.9 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 3.2 |
16.2 |
GO:0021965 |
spinal cord ventral commissure morphogenesis(GO:0021965) |
| 3.1 |
9.3 |
GO:0090425 |
hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 3.1 |
18.5 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 3.0 |
21.2 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 2.7 |
13.3 |
GO:0021856 |
cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 2.7 |
8.0 |
GO:0003195 |
tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) |
| 2.6 |
7.9 |
GO:0060067 |
cervix development(GO:0060067) |
| 2.4 |
19.3 |
GO:0097369 |
sodium ion import(GO:0097369) |
| 2.3 |
4.6 |
GO:0070885 |
negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 2.1 |
6.4 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 2.1 |
6.4 |
GO:2000686 |
regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 2.0 |
18.1 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 2.0 |
6.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 2.0 |
8.0 |
GO:0055099 |
regulation of Cdc42 protein signal transduction(GO:0032489) response to high density lipoprotein particle(GO:0055099) platelet dense granule organization(GO:0060155) |
| 1.8 |
25.8 |
GO:0021540 |
corpus callosum morphogenesis(GO:0021540) |
| 1.8 |
5.3 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 1.8 |
7.0 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 1.8 |
40.5 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 1.7 |
18.2 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 1.6 |
6.6 |
GO:0097393 |
post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.6 |
9.8 |
GO:0007258 |
JUN phosphorylation(GO:0007258) |
| 1.6 |
6.2 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 1.5 |
10.8 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 1.5 |
15.4 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.5 |
10.6 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 1.5 |
4.5 |
GO:0044849 |
estrous cycle(GO:0044849) |
| 1.5 |
1.5 |
GO:1900426 |
positive regulation of defense response to bacterium(GO:1900426) |
| 1.5 |
10.4 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 1.5 |
5.9 |
GO:0060083 |
smooth muscle contraction involved in micturition(GO:0060083) |
| 1.4 |
5.8 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 1.4 |
2.9 |
GO:2000790 |
regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 1.4 |
11.4 |
GO:0097090 |
presynaptic membrane organization(GO:0097090) |
| 1.4 |
5.6 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 1.4 |
8.3 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 1.4 |
13.8 |
GO:0033631 |
cell-cell adhesion mediated by integrin(GO:0033631) |
| 1.4 |
58.9 |
GO:2001222 |
regulation of neuron migration(GO:2001222) |
| 1.4 |
15.0 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 1.3 |
2.6 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 1.2 |
8.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 1.2 |
4.8 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 1.1 |
6.9 |
GO:0016198 |
axon choice point recognition(GO:0016198) |
| 1.1 |
11.4 |
GO:0033603 |
positive regulation of dopamine secretion(GO:0033603) |
| 1.1 |
2.2 |
GO:2000111 |
positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.1 |
3.3 |
GO:1900365 |
positive regulation of mRNA polyadenylation(GO:1900365) |
| 1.1 |
7.7 |
GO:0021796 |
cerebral cortex regionalization(GO:0021796) |
| 1.1 |
15.0 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 1.0 |
8.9 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 1.0 |
1.9 |
GO:0021812 |
neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 1.0 |
4.8 |
GO:0003105 |
negative regulation of glomerular filtration(GO:0003105) |
| 0.9 |
2.8 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) |
| 0.9 |
2.8 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 0.9 |
2.8 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.9 |
3.7 |
GO:0032289 |
central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 0.9 |
3.7 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.9 |
2.7 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.9 |
31.9 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.8 |
6.8 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.8 |
5.0 |
GO:2001181 |
positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.8 |
3.3 |
GO:0010808 |
positive regulation of synaptic vesicle priming(GO:0010808) positive regulation of synaptic vesicle exocytosis(GO:2000302) |
| 0.8 |
34.3 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.8 |
2.4 |
GO:1902528 |
regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.8 |
7.9 |
GO:0015721 |
bile acid and bile salt transport(GO:0015721) |
| 0.8 |
3.2 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.8 |
7.1 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.8 |
6.3 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.8 |
7.0 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.7 |
3.7 |
GO:0090310 |
negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.7 |
11.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.7 |
4.3 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.7 |
2.1 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.7 |
2.1 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.7 |
6.8 |
GO:0042572 |
retinol metabolic process(GO:0042572) |
| 0.7 |
2.0 |
GO:0010958 |
regulation of amino acid import(GO:0010958) regulation of L-arginine import(GO:0010963) |
| 0.7 |
6.7 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.7 |
15.3 |
GO:2000463 |
positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.7 |
2.0 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.6 |
3.9 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.6 |
2.6 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.6 |
1.3 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.6 |
52.7 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.6 |
3.8 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
3.8 |
GO:0001542 |
ovulation from ovarian follicle(GO:0001542) |
| 0.6 |
2.5 |
GO:0097039 |
protein linear polyubiquitination(GO:0097039) |
| 0.6 |
2.5 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.6 |
1.8 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.6 |
43.2 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.6 |
12.0 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.6 |
5.3 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.6 |
4.1 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.6 |
2.3 |
GO:1903587 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.6 |
1.7 |
GO:0042138 |
meiotic DNA double-strand break formation(GO:0042138) |
| 0.6 |
2.3 |
GO:0048520 |
positive regulation of behavior(GO:0048520) |
| 0.6 |
2.3 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) |
| 0.6 |
2.3 |
GO:0051012 |
microtubule sliding(GO:0051012) |
| 0.6 |
2.3 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.6 |
1.7 |
GO:0060753 |
regulation of mast cell chemotaxis(GO:0060753) |
| 0.5 |
6.6 |
GO:1904321 |
response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.5 |
11.7 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.5 |
1.1 |
GO:0060489 |
establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 |
2.1 |
GO:0002457 |
T cell antigen processing and presentation(GO:0002457) |
| 0.5 |
2.1 |
GO:0010756 |
positive regulation of plasminogen activation(GO:0010756) |
| 0.5 |
2.0 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.5 |
25.4 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.5 |
1.0 |
GO:0048341 |
paraxial mesoderm formation(GO:0048341) |
| 0.5 |
1.5 |
GO:0046958 |
nonassociative learning(GO:0046958) |
| 0.5 |
6.4 |
GO:0086064 |
cell communication by electrical coupling involved in cardiac conduction(GO:0086064) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.5 |
1.9 |
GO:0071442 |
regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.5 |
4.9 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.5 |
1.9 |
GO:0030397 |
membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.5 |
3.8 |
GO:1903421 |
regulation of synaptic vesicle recycling(GO:1903421) |
| 0.5 |
1.4 |
GO:0097167 |
circadian regulation of translation(GO:0097167) |
| 0.5 |
1.4 |
GO:0001580 |
detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) sensory perception of bitter taste(GO:0050913) |
| 0.5 |
3.3 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.5 |
39.9 |
GO:0007612 |
learning(GO:0007612) |
| 0.5 |
4.2 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.5 |
7.2 |
GO:2000114 |
regulation of establishment of cell polarity(GO:2000114) |
| 0.4 |
7.2 |
GO:0015693 |
magnesium ion transport(GO:0015693) |
| 0.4 |
1.3 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 0.4 |
1.7 |
GO:0035616 |
histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.4 |
1.7 |
GO:0050904 |
diapedesis(GO:0050904) |
| 0.4 |
1.7 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.4 |
2.1 |
GO:2001168 |
regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.4 |
1.7 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.4 |
4.6 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.4 |
4.6 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.4 |
17.2 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.4 |
2.9 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.4 |
2.8 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.4 |
1.2 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.4 |
1.6 |
GO:0070086 |
ubiquitin-dependent endocytosis(GO:0070086) |
| 0.4 |
2.0 |
GO:0046501 |
protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.4 |
1.2 |
GO:0019482 |
beta-alanine metabolic process(GO:0019482) |
| 0.4 |
3.1 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.4 |
1.5 |
GO:0015793 |
glycerol transport(GO:0015793) |
| 0.4 |
2.3 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.4 |
4.5 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.4 |
1.1 |
GO:0061588 |
calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.4 |
3.0 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.4 |
1.1 |
GO:1902950 |
regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.4 |
14.3 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.4 |
0.7 |
GO:2000664 |
positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.4 |
26.0 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.4 |
1.4 |
GO:0042088 |
T-helper 1 type immune response(GO:0042088) |
| 0.4 |
1.1 |
GO:0019858 |
cytosine metabolic process(GO:0019858) |
| 0.4 |
1.8 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.3 |
1.4 |
GO:0071360 |
cellular response to exogenous dsRNA(GO:0071360) |
| 0.3 |
2.1 |
GO:0015015 |
heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.3 |
2.0 |
GO:0070072 |
vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.3 |
1.7 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.3 |
2.0 |
GO:0046543 |
development of secondary female sexual characteristics(GO:0046543) |
| 0.3 |
5.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.3 |
1.0 |
GO:0042758 |
long-chain fatty acid catabolic process(GO:0042758) |
| 0.3 |
2.0 |
GO:0002524 |
hypersensitivity(GO:0002524) |
| 0.3 |
17.5 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.3 |
3.5 |
GO:0001504 |
neurotransmitter uptake(GO:0001504) |
| 0.3 |
4.0 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 |
5.2 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 |
1.5 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.3 |
0.9 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.3 |
0.9 |
GO:0050882 |
voluntary musculoskeletal movement(GO:0050882) |
| 0.3 |
2.4 |
GO:0090050 |
positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.3 |
4.2 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.3 |
0.6 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.3 |
11.3 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.3 |
1.4 |
GO:0030259 |
lipid glycosylation(GO:0030259) |
| 0.3 |
0.6 |
GO:0002578 |
negative regulation of antigen processing and presentation(GO:0002578) |
| 0.3 |
1.4 |
GO:0045654 |
positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.3 |
0.8 |
GO:0034454 |
microtubule anchoring at centrosome(GO:0034454) |
| 0.3 |
0.8 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) serine phosphorylation of STAT protein(GO:0042501) |
| 0.3 |
0.8 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 |
2.2 |
GO:0021859 |
pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.3 |
2.2 |
GO:0006691 |
leukotriene metabolic process(GO:0006691) |
| 0.3 |
2.2 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.3 |
6.7 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.3 |
1.3 |
GO:0031022 |
nuclear migration along microfilament(GO:0031022) |
| 0.3 |
0.5 |
GO:0014873 |
response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.3 |
1.9 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.3 |
3.7 |
GO:0097484 |
dendrite extension(GO:0097484) |
| 0.3 |
1.8 |
GO:0018344 |
protein geranylgeranylation(GO:0018344) |
| 0.3 |
8.9 |
GO:0048843 |
negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.3 |
1.6 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.3 |
9.0 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.2 |
0.7 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 |
2.4 |
GO:1903608 |
protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 |
2.6 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.2 |
4.1 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.2 |
1.6 |
GO:0007342 |
fusion of sperm to egg plasma membrane(GO:0007342) dendritic spine maintenance(GO:0097062) |
| 0.2 |
0.9 |
GO:0034727 |
lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) late nucleophagy(GO:0044805) single-organism membrane invagination(GO:1902534) |
| 0.2 |
2.0 |
GO:0010832 |
negative regulation of myotube differentiation(GO:0010832) |
| 0.2 |
1.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.2 |
0.7 |
GO:0070560 |
calcium-mediated signaling using extracellular calcium source(GO:0035585) protein secretion by platelet(GO:0070560) positive regulation of platelet aggregation(GO:1901731) |
| 0.2 |
0.4 |
GO:0055118 |
positive regulation of chronic inflammatory response(GO:0002678) negative regulation of cardiac muscle contraction(GO:0055118) |
| 0.2 |
0.9 |
GO:0046549 |
retinal cone cell development(GO:0046549) |
| 0.2 |
0.4 |
GO:0045410 |
positive regulation of interleukin-6 biosynthetic process(GO:0045410) negative regulation of branching morphogenesis of a nerve(GO:2000173) |
| 0.2 |
0.9 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.2 |
7.0 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.2 |
1.5 |
GO:0070102 |
interleukin-6-mediated signaling pathway(GO:0070102) |
| 0.2 |
2.1 |
GO:0042659 |
regulation of cell fate specification(GO:0042659) |
| 0.2 |
0.4 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.2 |
0.6 |
GO:0045085 |
negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.2 |
0.8 |
GO:1903726 |
negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.2 |
1.4 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.2 |
1.0 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 |
3.4 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.2 |
2.8 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.2 |
0.4 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.2 |
1.2 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.2 |
4.2 |
GO:0035335 |
peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 |
1.4 |
GO:0016584 |
nucleosome positioning(GO:0016584) |
| 0.2 |
1.6 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
1.6 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.2 |
2.9 |
GO:0042759 |
long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 |
7.2 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.2 |
0.8 |
GO:0071105 |
macrophage activation involved in immune response(GO:0002281) response to interleukin-11(GO:0071105) osteoclast fusion(GO:0072675) |
| 0.2 |
2.3 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 |
3.9 |
GO:2000060 |
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.2 |
1.3 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.2 |
0.8 |
GO:0006848 |
pyruvate transport(GO:0006848) |
| 0.2 |
3.8 |
GO:0034612 |
response to tumor necrosis factor(GO:0034612) |
| 0.2 |
0.9 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.2 |
2.8 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 |
19.0 |
GO:0071805 |
potassium ion transmembrane transport(GO:0071805) |
| 0.2 |
0.7 |
GO:0090399 |
replicative senescence(GO:0090399) |
| 0.2 |
0.7 |
GO:0033540 |
fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.2 |
1.1 |
GO:0035428 |
hexose transmembrane transport(GO:0035428) |
| 0.2 |
1.7 |
GO:0060049 |
regulation of protein glycosylation(GO:0060049) |
| 0.2 |
1.2 |
GO:0006020 |
inositol metabolic process(GO:0006020) |
| 0.2 |
15.8 |
GO:0045727 |
positive regulation of translation(GO:0045727) |
| 0.2 |
5.5 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.2 |
0.3 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 |
1.5 |
GO:0051764 |
actin crosslink formation(GO:0051764) |
| 0.2 |
0.3 |
GO:0060467 |
negative regulation of fertilization(GO:0060467) |
| 0.2 |
2.0 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.2 |
1.0 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 |
6.7 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.2 |
0.6 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.2 |
1.7 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.2 |
3.8 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.1 |
3.0 |
GO:0048535 |
lymph node development(GO:0048535) |
| 0.1 |
0.4 |
GO:0015712 |
hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 |
0.4 |
GO:0098908 |
regulation of neuronal action potential(GO:0098908) positive regulation of neuronal action potential(GO:1904457) |
| 0.1 |
0.6 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.1 |
0.3 |
GO:2000418 |
positive regulation of eosinophil migration(GO:2000418) |
| 0.1 |
1.7 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.1 |
1.7 |
GO:0071392 |
cellular response to estradiol stimulus(GO:0071392) |
| 0.1 |
0.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.1 |
3.2 |
GO:1900026 |
positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 |
0.7 |
GO:0060340 |
positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.1 |
1.5 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.1 |
0.5 |
GO:0019336 |
phenol-containing compound catabolic process(GO:0019336) |
| 0.1 |
2.0 |
GO:0051016 |
barbed-end actin filament capping(GO:0051016) |
| 0.1 |
0.9 |
GO:2000628 |
regulation of miRNA metabolic process(GO:2000628) |
| 0.1 |
0.3 |
GO:0042939 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 |
0.9 |
GO:0042126 |
nitrate metabolic process(GO:0042126) |
| 0.1 |
5.9 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
0.6 |
GO:0045176 |
apical protein localization(GO:0045176) |
| 0.1 |
0.4 |
GO:0031133 |
peripheral nervous system axon regeneration(GO:0014012) regulation of axon diameter(GO:0031133) intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.2 |
GO:0010624 |
regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 |
1.0 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 |
7.7 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.1 |
1.4 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.1 |
1.0 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.1 |
0.6 |
GO:0060164 |
regulation of timing of neuron differentiation(GO:0060164) |
| 0.1 |
1.6 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
0.4 |
GO:0045745 |
positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 |
1.0 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.1 |
GO:0030813 |
positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.1 |
0.9 |
GO:0035608 |
protein deglutamylation(GO:0035608) |
| 0.1 |
0.6 |
GO:0061635 |
regulation of protein complex stability(GO:0061635) |
| 0.1 |
0.3 |
GO:0006420 |
arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 |
0.3 |
GO:0002677 |
regulation of chronic inflammatory response(GO:0002676) negative regulation of chronic inflammatory response(GO:0002677) |
| 0.1 |
0.8 |
GO:0007175 |
negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 |
0.6 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
0.5 |
GO:0031293 |
membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 |
1.0 |
GO:0006497 |
protein lipidation(GO:0006497) |
| 0.1 |
0.2 |
GO:1900042 |
positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 |
0.6 |
GO:0050807 |
regulation of synapse organization(GO:0050807) |
| 0.1 |
0.4 |
GO:0010796 |
regulation of multivesicular body size(GO:0010796) |
| 0.1 |
0.4 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.1 |
0.9 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
1.1 |
GO:0060117 |
auditory receptor cell development(GO:0060117) |
| 0.1 |
0.1 |
GO:1901492 |
positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 |
0.4 |
GO:0060179 |
male mating behavior(GO:0060179) |
| 0.1 |
1.6 |
GO:0048168 |
regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.1 |
0.4 |
GO:0010735 |
positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 |
1.4 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 |
1.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 |
0.5 |
GO:0045162 |
neuronal ion channel clustering(GO:0045161) clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 |
1.0 |
GO:0060213 |
regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.1 |
0.7 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.4 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
1.7 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.1 |
0.4 |
GO:1902414 |
protein localization to cell junction(GO:1902414) |
| 0.1 |
1.3 |
GO:0006828 |
manganese ion transport(GO:0006828) |
| 0.1 |
1.5 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.1 |
4.3 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.1 |
1.1 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 |
0.5 |
GO:0060546 |
negative regulation of necroptotic process(GO:0060546) |
| 0.1 |
4.3 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 |
0.8 |
GO:0001881 |
receptor recycling(GO:0001881) |
| 0.1 |
0.5 |
GO:0010528 |
regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.1 |
1.2 |
GO:0015813 |
L-glutamate transport(GO:0015813) |
| 0.1 |
0.8 |
GO:2000096 |
positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 |
0.5 |
GO:0032486 |
Rap protein signal transduction(GO:0032486) |
| 0.1 |
0.2 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.1 |
2.4 |
GO:0006865 |
amino acid transport(GO:0006865) |
| 0.1 |
0.4 |
GO:0072429 |
response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 |
0.4 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.1 |
0.5 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.1 |
0.5 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.2 |
GO:0070213 |
protein auto-ADP-ribosylation(GO:0070213) |
| 0.1 |
0.2 |
GO:0030321 |
transepithelial chloride transport(GO:0030321) |
| 0.1 |
0.6 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
1.4 |
GO:0008333 |
endosome to lysosome transport(GO:0008333) |
| 0.1 |
5.4 |
GO:0008286 |
insulin receptor signaling pathway(GO:0008286) |
| 0.1 |
1.2 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.1 |
0.5 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.1 |
0.6 |
GO:0006857 |
oligopeptide transport(GO:0006857) |
| 0.1 |
7.0 |
GO:0006333 |
chromatin assembly or disassembly(GO:0006333) |
| 0.1 |
0.1 |
GO:0038161 |
prolactin signaling pathway(GO:0038161) |
| 0.1 |
0.5 |
GO:0006953 |
acute-phase response(GO:0006953) |
| 0.1 |
1.7 |
GO:0031110 |
regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 |
1.3 |
GO:0035458 |
cellular response to interferon-beta(GO:0035458) |
| 0.1 |
1.4 |
GO:0046488 |
phosphatidylinositol metabolic process(GO:0046488) |
| 0.1 |
0.2 |
GO:0008594 |
photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 |
0.3 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.1 |
2.2 |
GO:0006891 |
intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 |
0.9 |
GO:0008053 |
mitochondrial fusion(GO:0008053) |
| 0.1 |
1.3 |
GO:0072583 |
clathrin-mediated endocytosis(GO:0072583) |
| 0.1 |
0.2 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.5 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.5 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.0 |
0.5 |
GO:0099500 |
synaptic vesicle fusion to presynaptic active zone membrane(GO:0031629) vesicle fusion to plasma membrane(GO:0099500) |
| 0.0 |
0.6 |
GO:0002920 |
regulation of humoral immune response(GO:0002920) |
| 0.0 |
0.7 |
GO:0032094 |
response to food(GO:0032094) |
| 0.0 |
4.0 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.0 |
0.6 |
GO:0030033 |
microvillus assembly(GO:0030033) |
| 0.0 |
0.2 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.8 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
1.0 |
GO:0045071 |
negative regulation of viral genome replication(GO:0045071) |
| 0.0 |
1.3 |
GO:0070979 |
protein K11-linked ubiquitination(GO:0070979) |
| 0.0 |
0.4 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.8 |
GO:0070536 |
protein K63-linked deubiquitination(GO:0070536) |
| 0.0 |
0.5 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.0 |
0.2 |
GO:1901030 |
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.0 |
1.2 |
GO:0006414 |
translational elongation(GO:0006414) |
| 0.0 |
0.2 |
GO:0061718 |
NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glycolytic process through glucose-1-phosphate(GO:0061622) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 |
0.2 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.0 |
0.5 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.9 |
GO:0006813 |
potassium ion transport(GO:0006813) |
| 0.0 |
0.3 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.1 |
GO:0071218 |
cellular response to misfolded protein(GO:0071218) |
| 0.0 |
0.3 |
GO:0015838 |
amino-acid betaine transport(GO:0015838) |
| 0.0 |
0.1 |
GO:0045822 |
negative regulation of heart contraction(GO:0045822) |
| 0.0 |
0.8 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 |
0.3 |
GO:0032464 |
positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 |
0.1 |
GO:0090209 |
negative regulation of triglyceride metabolic process(GO:0090209) |
| 0.0 |
0.8 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
1.6 |
GO:0006367 |
transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 |
1.3 |
GO:0007611 |
learning or memory(GO:0007611) |
| 0.0 |
0.3 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.4 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.0 |
0.1 |
GO:0019276 |
UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 |
1.5 |
GO:0021549 |
cerebellum development(GO:0021549) |
| 0.0 |
1.0 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.5 |
GO:0001945 |
lymph vessel development(GO:0001945) |
| 0.0 |
0.1 |
GO:1901623 |
regulation of lymphocyte chemotaxis(GO:1901623) |
| 0.0 |
1.7 |
GO:0090630 |
activation of GTPase activity(GO:0090630) |
| 0.0 |
0.3 |
GO:0014887 |
cardiac muscle adaptation(GO:0014887) |
| 0.0 |
0.2 |
GO:0070301 |
cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 |
0.4 |
GO:0042742 |
defense response to bacterium(GO:0042742) |
| 0.0 |
0.3 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 |
2.7 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.1 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.0 |
0.4 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.2 |
GO:0031274 |
positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 |
0.3 |
GO:0044804 |
nucleophagy(GO:0044804) |
| 0.0 |
0.4 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.2 |
GO:0050667 |
homocysteine metabolic process(GO:0050667) |
| 0.0 |
0.0 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
1.9 |
GO:0007018 |
microtubule-based movement(GO:0007018) |
| 0.0 |
0.1 |
GO:0033004 |
negative regulation of mast cell activation(GO:0033004) |
| 0.0 |
0.3 |
GO:0035518 |
histone H2A monoubiquitination(GO:0035518) |
| 0.0 |
0.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.1 |
GO:2001197 |
regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.0 |
3.0 |
GO:0006914 |
autophagy(GO:0006914) |