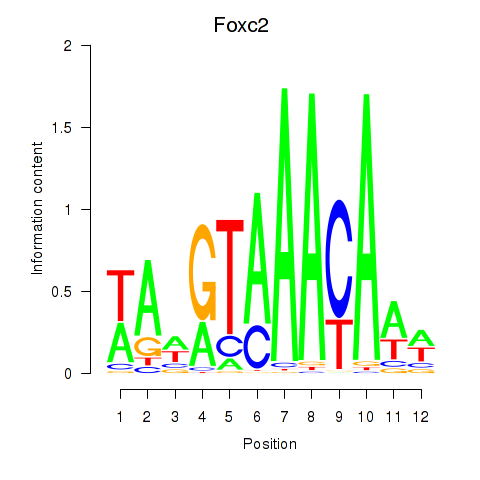
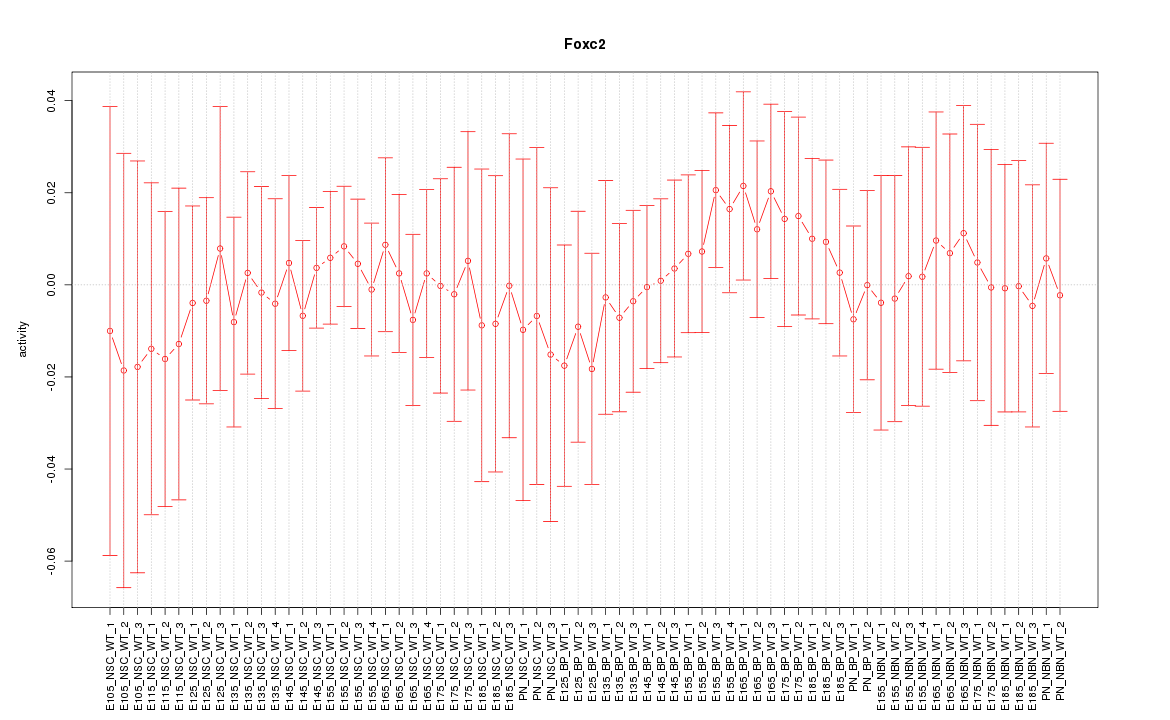
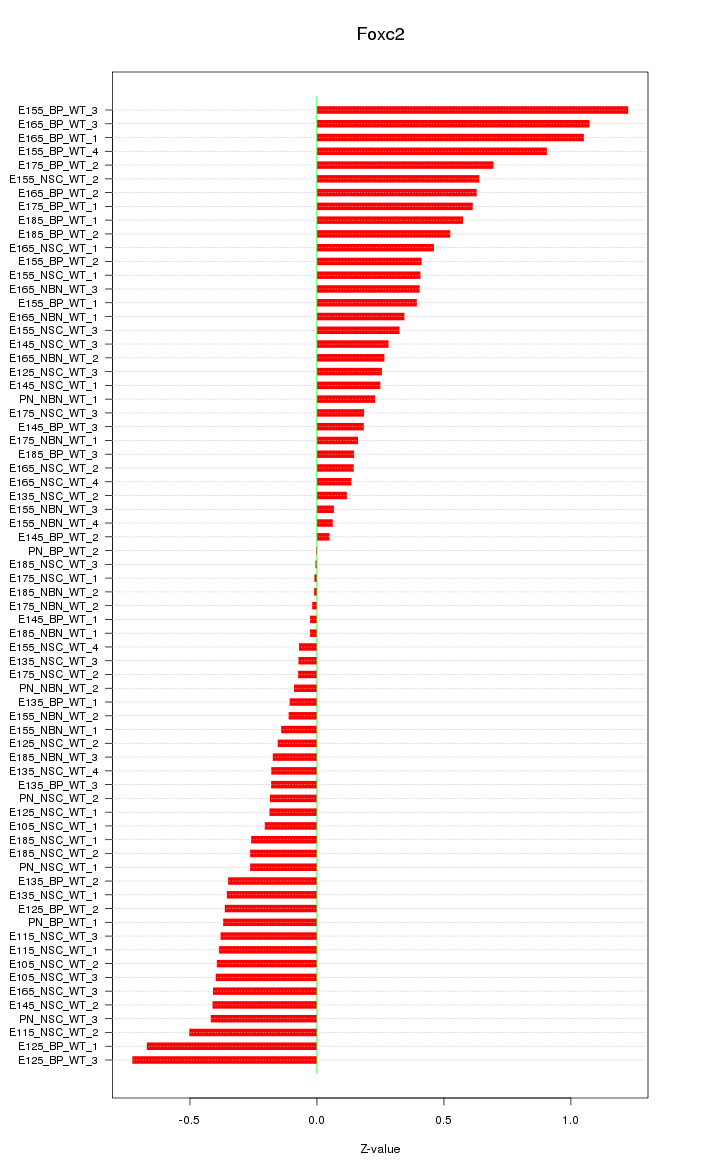
| 1.1 |
3.4 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.0 |
4.0 |
GO:1901894 |
regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.8 |
2.3 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 0.5 |
1.5 |
GO:0042998 |
positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.5 |
3.4 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.5 |
4.7 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.5 |
6.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.4 |
2.2 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.3 |
1.4 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.3 |
0.8 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.3 |
1.6 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.2 |
0.7 |
GO:0007525 |
somatic muscle development(GO:0007525) |
| 0.2 |
0.9 |
GO:1903898 |
positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.2 |
4.5 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.2 |
1.0 |
GO:0036438 |
maintenance of lens transparency(GO:0036438) |
| 0.2 |
1.0 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.2 |
1.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.2 |
0.6 |
GO:0010637 |
negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 |
2.5 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
1.5 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.1 |
1.9 |
GO:0070842 |
aggresome assembly(GO:0070842) |
| 0.1 |
0.6 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 |
0.5 |
GO:0042117 |
monocyte activation(GO:0042117) |
| 0.1 |
1.1 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.4 |
GO:0008295 |
spermidine biosynthetic process(GO:0008295) |
| 0.1 |
0.6 |
GO:0001927 |
exocyst assembly(GO:0001927) |
| 0.1 |
0.4 |
GO:0072344 |
rescue of stalled ribosome(GO:0072344) |
| 0.1 |
0.8 |
GO:0006390 |
transcription from mitochondrial promoter(GO:0006390) |
| 0.1 |
0.4 |
GO:0090164 |
asymmetric Golgi ribbon formation(GO:0090164) |
| 0.1 |
0.6 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.1 |
0.7 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.4 |
GO:0002317 |
plasma cell differentiation(GO:0002317) |
| 0.1 |
1.1 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 |
0.3 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 |
2.2 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.1 |
0.3 |
GO:1903599 |
positive regulation of mitophagy(GO:1903599) |
| 0.1 |
0.5 |
GO:0010748 |
negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.1 |
0.8 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.2 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.1 |
0.6 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.1 |
0.8 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.1 |
1.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 |
0.3 |
GO:0018243 |
protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 |
0.3 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.0 |
0.4 |
GO:0060613 |
fat pad development(GO:0060613) |
| 0.0 |
2.9 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.0 |
1.1 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.0 |
1.4 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.0 |
0.2 |
GO:0072015 |
glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 |
2.1 |
GO:0043268 |
positive regulation of potassium ion transport(GO:0043268) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.1 |
GO:0038145 |
macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 |
1.3 |
GO:0036465 |
synaptic vesicle recycling(GO:0036465) |
| 0.0 |
1.1 |
GO:0050919 |
negative chemotaxis(GO:0050919) |
| 0.0 |
0.2 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.0 |
0.4 |
GO:0001553 |
luteinization(GO:0001553) |
| 0.0 |
0.8 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.5 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 |
0.1 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
0.6 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.0 |
0.2 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0005981 |
regulation of glycogen catabolic process(GO:0005981) |
| 0.0 |
0.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 |
0.3 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.0 |
0.9 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |