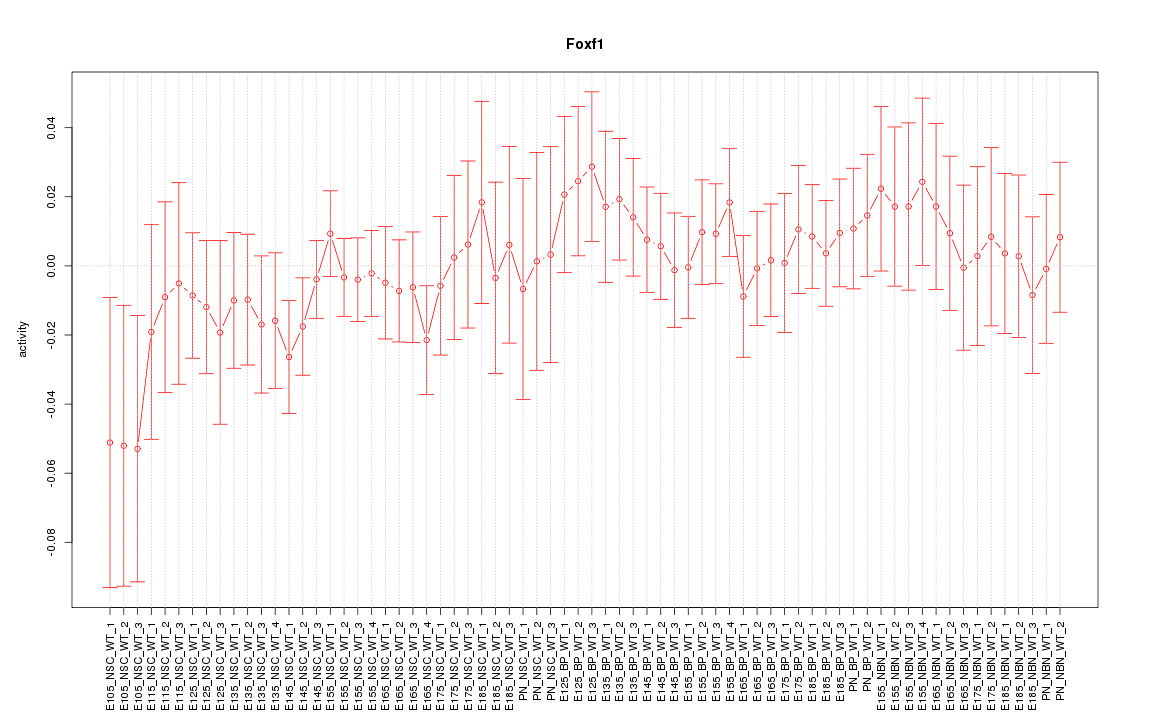
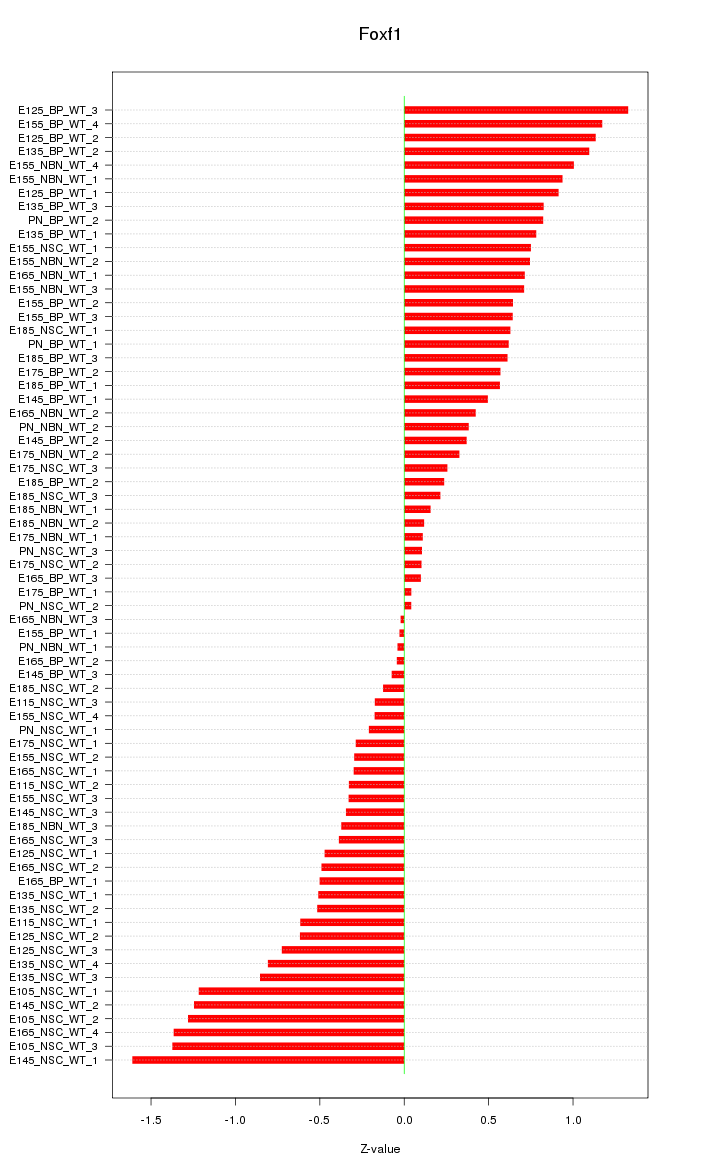
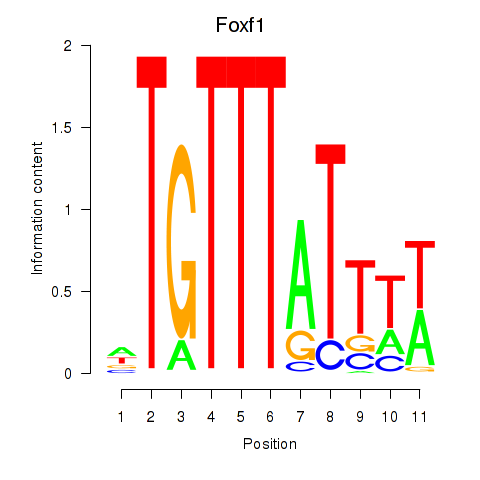
Motif ID: Foxf1
Z-value: 0.677
Transcription factors associated with Foxf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Foxf1 | ENSMUSG00000042812.4 | Foxf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxf1 | mm10_v2_chr8_+_121084352_121084474 | -0.29 | 1.6e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 12.4 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.2 | 3.5 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 1.1 | 4.3 | GO:0032512 | regulation of protein phosphatase type 2B activity(GO:0032512) positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 1.0 | 3.0 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.9 | 3.6 | GO:0008627 | intrinsic apoptotic signaling pathway in response to osmotic stress(GO:0008627) |
| 0.8 | 2.5 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.8 | 10.7 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.7 | 2.2 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.7 | 2.0 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 | 8.9 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.6 | 1.7 | GO:0051464 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) positive regulation of cortisol secretion(GO:0051464) |
| 0.6 | 2.2 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 0.5 | 1.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.5 | 1.5 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.5 | 1.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.5 | 10.7 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.5 | 4.8 | GO:0007379 | segment specification(GO:0007379) |
| 0.5 | 2.8 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.4 | 1.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.4 | 2.6 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.4 | 1.3 | GO:0046881 | sperm ejaculation(GO:0042713) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.4 | 2.5 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 | 5.4 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.4 | 3.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.4 | 8.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.4 | 3.4 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 3.0 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.3 | 2.3 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.3 | 3.0 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.3 | 5.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.3 | 2.0 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.3 | 1.4 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.3 | 2.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 | 5.1 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.3 | 2.5 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.2 | 0.9 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.8 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.2 | 1.0 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.2 | 2.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.2 | 2.3 | GO:1902018 | negative regulation of cilium assembly(GO:1902018) |
| 0.2 | 2.6 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 0.6 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.2 | 0.7 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 1.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.2 | 1.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 2.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 2.4 | GO:1902260 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.2 | 5.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.2 | 0.5 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 0.2 | 0.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) regulation of dendritic spine maintenance(GO:1902950) positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.1 | 0.4 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 3.3 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 0.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.7 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.1 | 0.8 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 1.1 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.4 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.9 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.1 | 4.4 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.1 | 0.8 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.1 | 2.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.9 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 0.8 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.1 | 0.7 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.7 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.7 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 3.5 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.1 | 2.7 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 1.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 5.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 2.1 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 0.6 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 | 0.9 | GO:2000052 | positive regulation of non-canonical Wnt signaling pathway(GO:2000052) |
| 0.1 | 4.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.6 | GO:0021766 | hippocampus development(GO:0021766) |
| 0.1 | 5.3 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 0.3 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 3.9 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 1.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.3 | GO:0072176 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 3.0 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 | 0.8 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.4 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.6 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.0 | 1.4 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.4 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 1.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.4 | GO:0002021 | response to dietary excess(GO:0002021) response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.5 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 1.0 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 2.9 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 2.4 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.1 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.3 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 1.3 | GO:0098792 | xenophagy(GO:0098792) |
| 0.0 | 0.3 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.1 | GO:0007603 | phototransduction, visible light(GO:0007603) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 8.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.7 | 14.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.6 | 5.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 1.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.4 | 3.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.5 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.4 | 3.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.3 | 6.5 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 2.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 1.5 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.3 | 3.9 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.2 | 0.7 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.2 | 3.9 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.2 | 3.6 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 0.6 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 0.8 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.1 | 1.0 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 2.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 5.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 2.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 5.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 2.0 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 4.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.2 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 2.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 3.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.5 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.1 | 4.4 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 16.2 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 2.2 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.0 | 13.3 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.0 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 2.5 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.8 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 2.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 1.5 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.6 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 6.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.4 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.8 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 2.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.4 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.1 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 5.1 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 1.4 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 10.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 1.3 | 10.7 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 1.0 | 3.0 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.9 | 12.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.8 | 2.5 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.7 | 2.2 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.7 | 5.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.7 | 5.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.6 | 3.5 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.5 | 4.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 1.5 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 0.3 | 0.7 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.3 | 2.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.3 | 4.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.3 | 2.0 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 7.3 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.3 | 1.5 | GO:0071253 | connexin binding(GO:0071253) |
| 0.3 | 1.3 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.2 | 1.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.2 | 3.9 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 2.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.2 | 4.7 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.2 | 0.7 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.2 | 2.0 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.2 | 0.8 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.2 | 2.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.2 | 2.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 1.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 4.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 3.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 4.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 1.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.1 | 2.0 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 1.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.8 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.1 | 3.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.8 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 2.5 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 0.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 0.8 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.1 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 2.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 2.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 2.0 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.5 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.0 | 0.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 3.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.4 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 2.5 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 2.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 1.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 1.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.9 | GO:0051087 | chaperone binding(GO:0051087) |