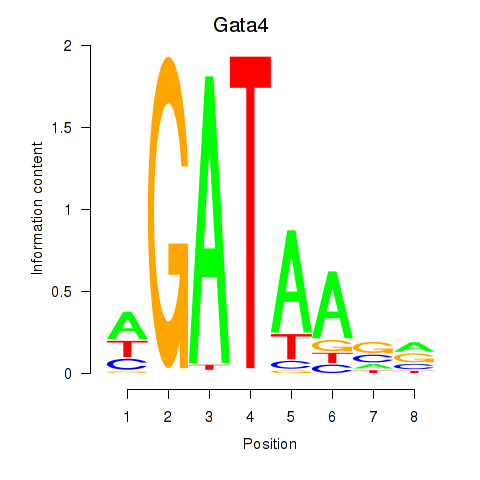
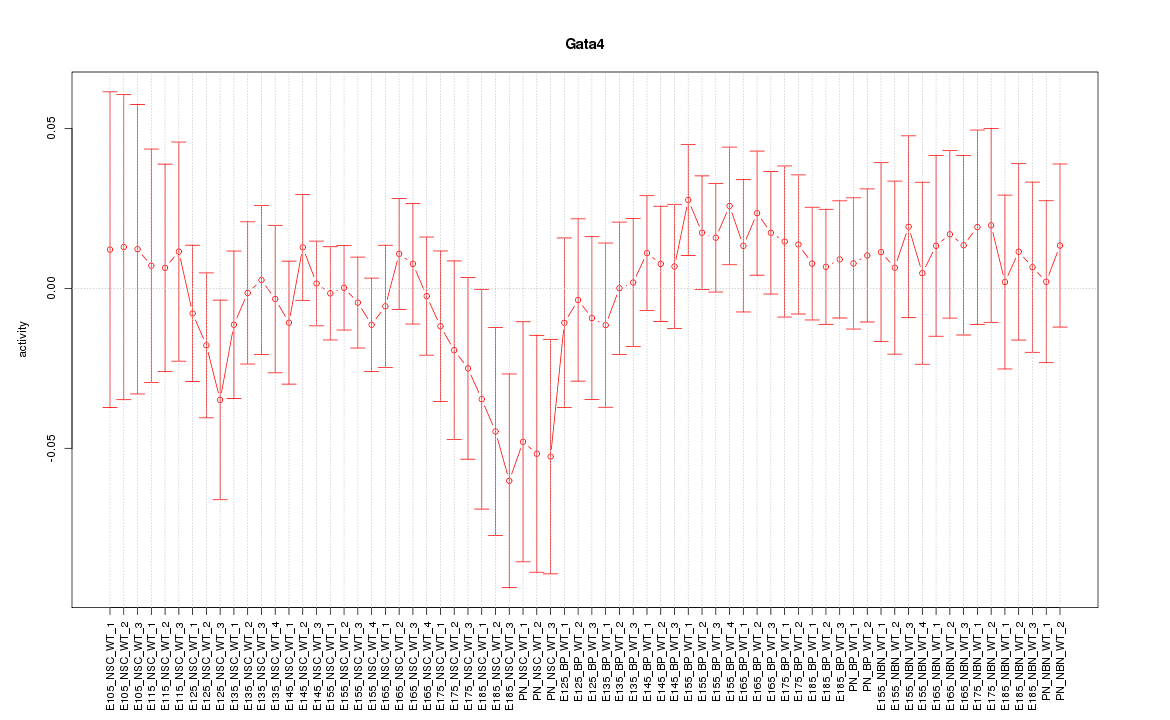
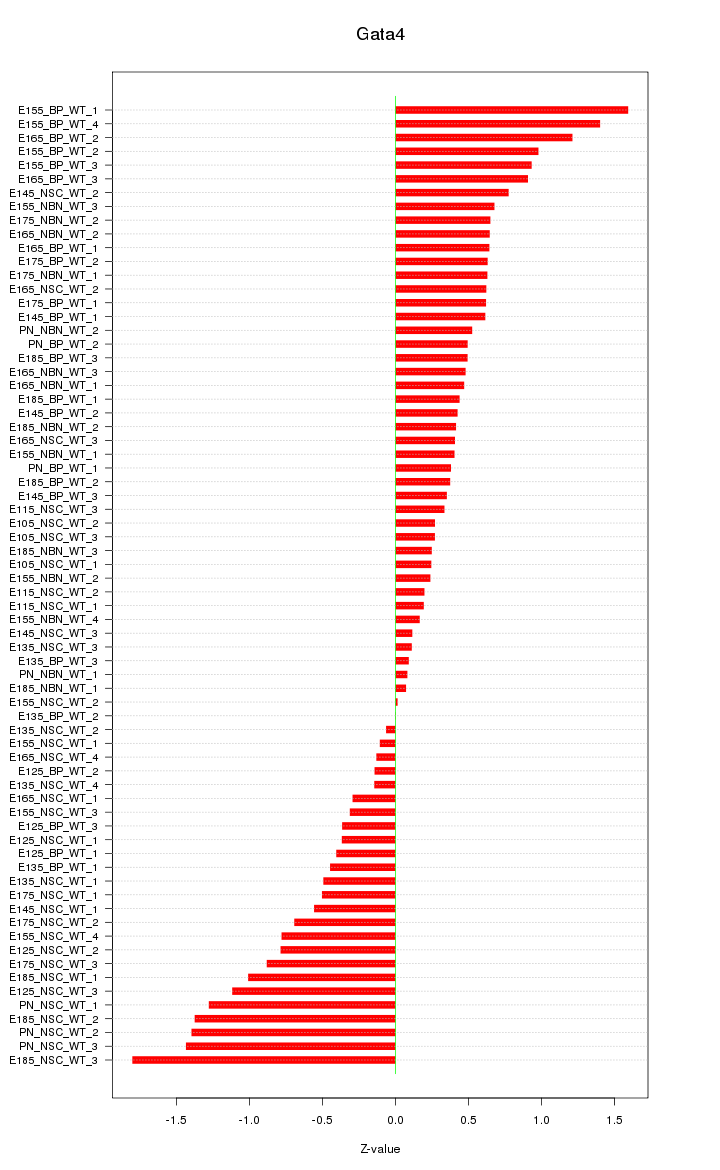
| 1.5 |
7.4 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 1.2 |
9.7 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 1.0 |
5.1 |
GO:0060414 |
aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 |
3.8 |
GO:0010359 |
regulation of anion channel activity(GO:0010359) |
| 0.6 |
9.7 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 |
1.8 |
GO:0097155 |
fasciculation of sensory neuron axon(GO:0097155) |
| 0.6 |
1.7 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 0.6 |
5.8 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.5 |
2.4 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.4 |
1.3 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.4 |
1.3 |
GO:0030300 |
regulation of intestinal cholesterol absorption(GO:0030300) |
| 0.4 |
1.2 |
GO:0060214 |
stem cell fate commitment(GO:0048865) stem cell fate specification(GO:0048866) endocardium formation(GO:0060214) cardiac cell fate determination(GO:0060913) regulation of cardiac cell fate specification(GO:2000043) |
| 0.4 |
3.0 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.3 |
1.0 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.3 |
12.0 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.3 |
0.9 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.3 |
1.2 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.3 |
2.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.3 |
2.0 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.3 |
1.9 |
GO:0006071 |
glycerol metabolic process(GO:0006071) |
| 0.3 |
0.8 |
GO:0070428 |
granuloma formation(GO:0002432) negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.3 |
1.6 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.3 |
1.0 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.2 |
2.2 |
GO:0050957 |
equilibrioception(GO:0050957) |
| 0.2 |
2.0 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.2 |
0.7 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.2 |
1.1 |
GO:0007262 |
STAT protein import into nucleus(GO:0007262) positive regulation of hyaluronan biosynthetic process(GO:1900127) |
| 0.2 |
1.3 |
GO:0042866 |
pyruvate biosynthetic process(GO:0042866) |
| 0.2 |
1.5 |
GO:0015862 |
uridine transport(GO:0015862) |
| 0.2 |
1.0 |
GO:0030091 |
protein repair(GO:0030091) |
| 0.2 |
0.6 |
GO:0030327 |
prenylated protein catabolic process(GO:0030327) |
| 0.2 |
1.0 |
GO:0032911 |
nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 |
3.2 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.2 |
3.0 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.2 |
0.6 |
GO:0002430 |
complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 |
5.9 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.2 |
0.8 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 |
1.1 |
GO:0061343 |
cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.2 |
2.0 |
GO:0097067 |
negative regulation of erythrocyte differentiation(GO:0045647) cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.2 |
2.3 |
GO:0042541 |
hemoglobin biosynthetic process(GO:0042541) |
| 0.2 |
3.6 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.1 |
1.0 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
0.9 |
GO:0001955 |
blood vessel maturation(GO:0001955) |
| 0.1 |
1.1 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
1.2 |
GO:0046485 |
ether lipid metabolic process(GO:0046485) |
| 0.1 |
7.3 |
GO:0045599 |
negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 |
1.0 |
GO:0086023 |
adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 |
1.9 |
GO:0021702 |
cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 |
0.5 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.1 |
4.0 |
GO:0035640 |
exploration behavior(GO:0035640) |
| 0.1 |
4.4 |
GO:0001541 |
ovarian follicle development(GO:0001541) |
| 0.1 |
0.6 |
GO:0002002 |
regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 |
6.0 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.1 |
2.9 |
GO:0090162 |
establishment of epithelial cell polarity(GO:0090162) |
| 0.1 |
1.1 |
GO:0046597 |
negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 |
2.1 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.1 |
3.6 |
GO:1902653 |
cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 |
4.0 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.1 |
0.7 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 |
0.5 |
GO:0030035 |
microspike assembly(GO:0030035) |
| 0.1 |
5.2 |
GO:0031532 |
actin cytoskeleton reorganization(GO:0031532) |
| 0.1 |
2.5 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
1.1 |
GO:0045197 |
establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 |
1.1 |
GO:0030970 |
retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 |
2.4 |
GO:0006096 |
glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 |
0.6 |
GO:0019216 |
regulation of lipid metabolic process(GO:0019216) |
| 0.0 |
0.1 |
GO:1904995 |
negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 |
0.1 |
GO:0036444 |
calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 |
0.2 |
GO:0045332 |
phospholipid translocation(GO:0045332) |
| 0.0 |
0.3 |
GO:0070935 |
3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 |
0.0 |
GO:1904139 |
microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 |
0.5 |
GO:0031114 |
negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 |
0.1 |
GO:0032693 |
negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 |
3.6 |
GO:0051056 |
regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 |
0.0 |
GO:0060686 |
regulation of prostatic bud formation(GO:0060685) negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 |
0.4 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |