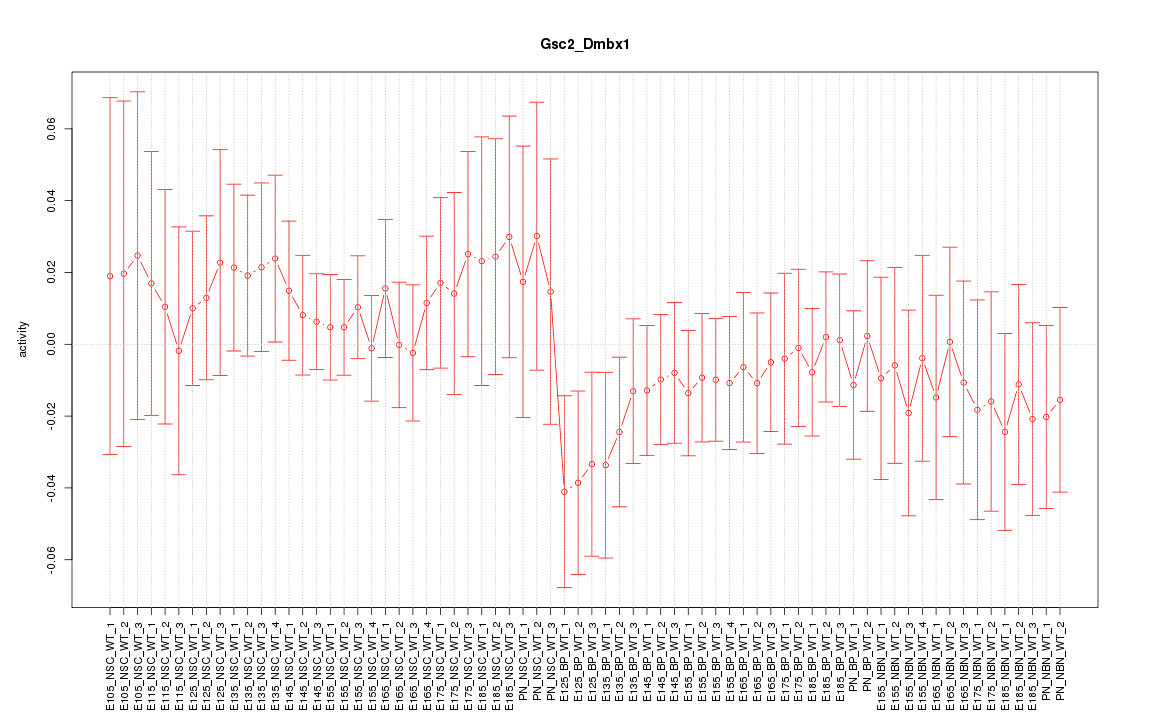
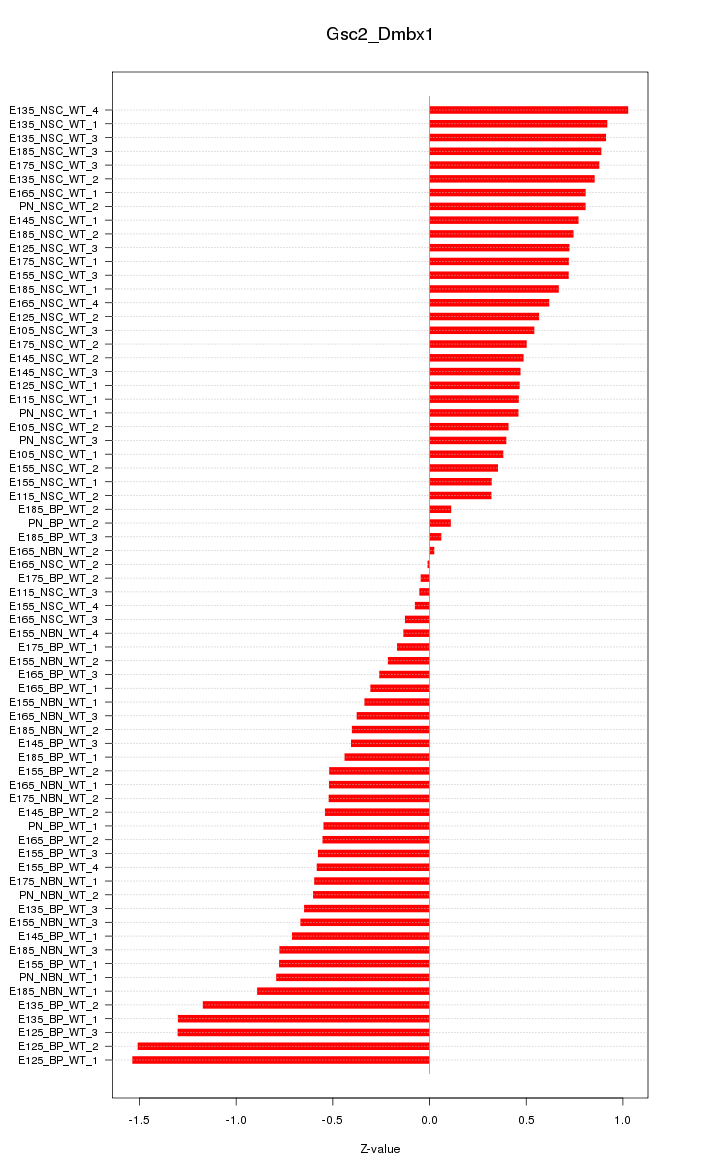
Motif ID: Gsc2_Dmbx1
Z-value: 0.657
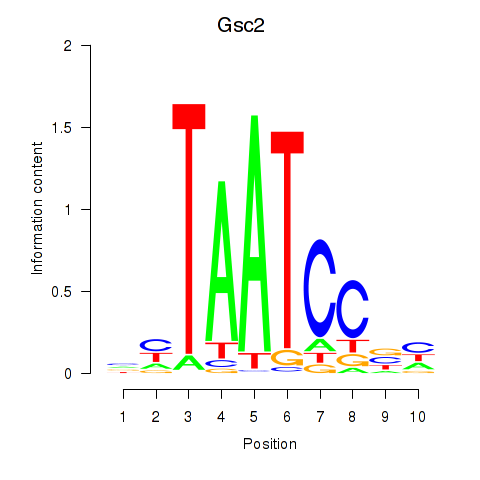
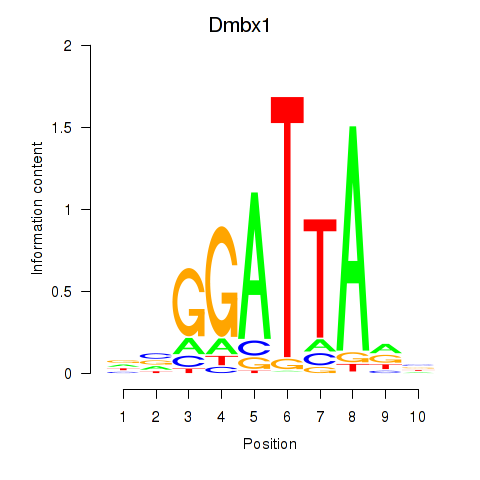
Transcription factors associated with Gsc2_Dmbx1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dmbx1 | ENSMUSG00000028707.9 | Dmbx1 |
| Gsc2 | ENSMUSG00000022738.6 | Gsc2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dmbx1 | mm10_v2_chr4_-_115939923_115939928 | 0.36 | 2.3e-03 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 2.2 | 8.8 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 1.5 | 8.7 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 1.3 | 3.8 | GO:1902524 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) negative regulation of interferon-beta biosynthetic process(GO:0045358) positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 1.0 | 2.9 | GO:0032650 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) cardiac cell fate determination(GO:0060913) |
| 0.9 | 4.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.9 | 2.6 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.8 | 3.2 | GO:2000705 | regulation of dense core granule biogenesis(GO:2000705) |
| 0.7 | 5.7 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.5 | 15.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.5 | 4.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.4 | 1.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.4 | 1.7 | GO:0046533 | negative regulation of photoreceptor cell differentiation(GO:0046533) |
| 0.4 | 1.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.3 | 7.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.3 | 1.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 | 1.6 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.2 | 3.0 | GO:2000010 | positive regulation of protein localization to cell surface(GO:2000010) |
| 0.2 | 2.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.6 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 5.8 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 1.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.4 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.1 | 1.1 | GO:0090331 | negative regulation of platelet aggregation(GO:0090331) |
| 0.1 | 2.6 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 1.9 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 2.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.3 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.1 | 1.6 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.1 | 0.3 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.9 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 1.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.8 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 1.7 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.7 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 1.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.8 | GO:0000910 | cytokinesis(GO:0000910) |
| 0.0 | 0.9 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 15.7 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.6 | 1.7 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.5 | 7.8 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.3 | 8.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 12.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.2 | 2.6 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.1 | 5.7 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 2.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 0.8 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.1 | 2.6 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.6 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.1 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 3.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 3.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 1.3 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0005839 | proteasome core complex(GO:0005839) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.9 | 15.7 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.8 | 4.7 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.6 | 8.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.5 | 4.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.5 | 7.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.5 | 1.9 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.4 | 1.6 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.3 | 12.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.3 | 1.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 2.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.3 | 3.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 1.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.2 | 1.0 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.1 | 3.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 0.4 | GO:0070140 | isopeptidase activity(GO:0070122) ubiquitin-like protein-specific isopeptidase activity(GO:0070138) SUMO-specific isopeptidase activity(GO:0070140) |
| 0.1 | 1.6 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.1 | 3.6 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 1.7 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 2.9 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.1 | 0.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 14.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.1 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 2.0 | GO:0042974 | retinoic acid receptor binding(GO:0042974) |
| 0.0 | 2.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 1.9 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 2.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.5 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.0 | 0.9 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |