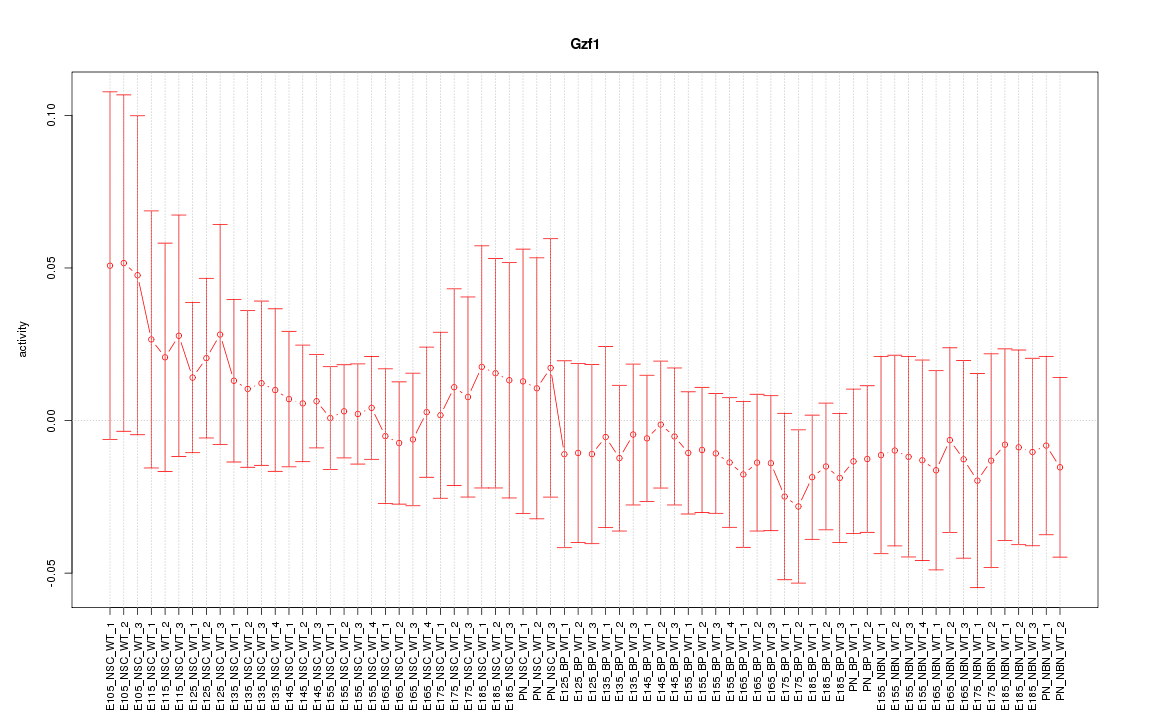
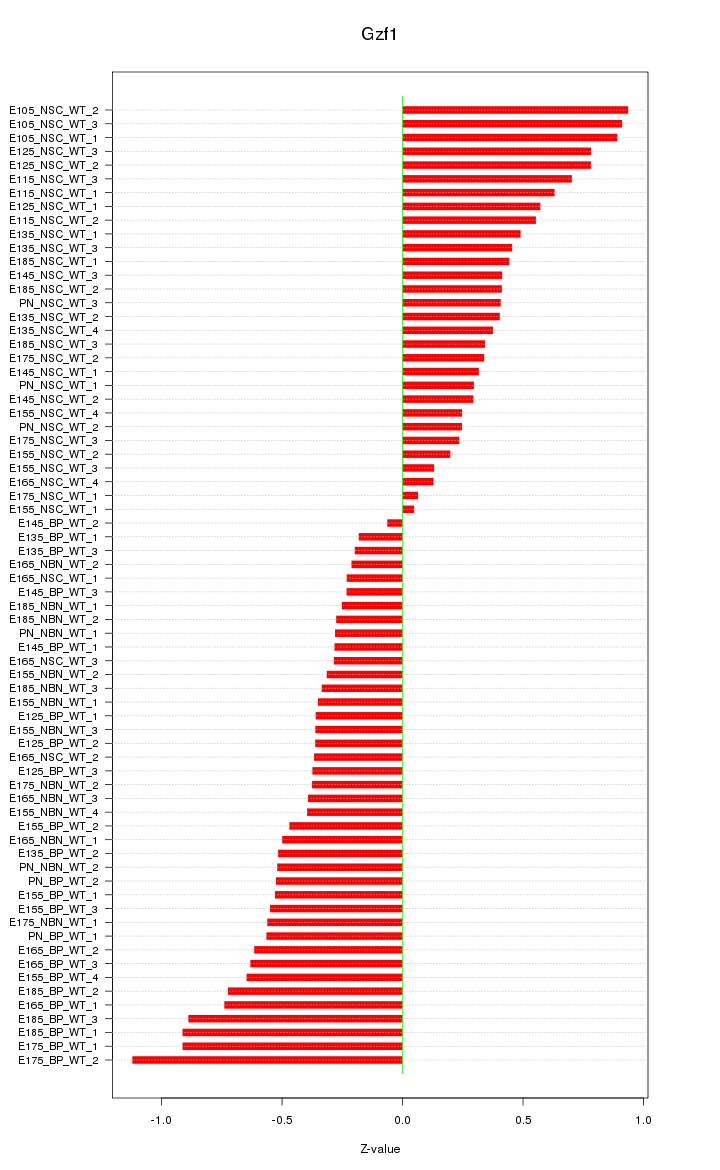
Motif ID: Gzf1
Z-value: 0.507
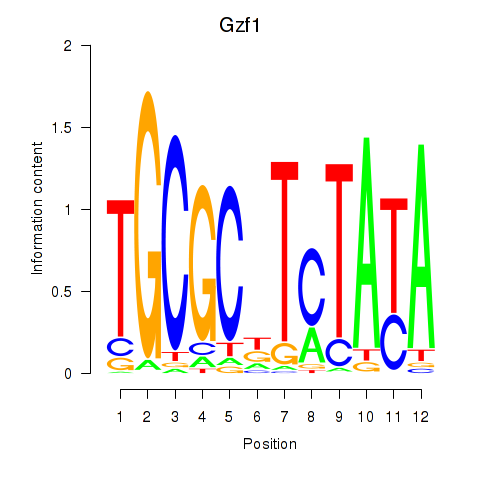
Transcription factors associated with Gzf1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Gzf1 | ENSMUSG00000027439.9 | Gzf1 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Gzf1 | mm10_v2_chr2_+_148681023_148681197 | -0.12 | 3.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.2 | 12.7 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.9 | 3.7 | GO:1904749 | rRNA export from nucleus(GO:0006407) regulation of protein localization to nucleolus(GO:1904749) positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.5 | 1.5 | GO:0031627 | telomeric loop formation(GO:0031627) |
| 0.5 | 4.1 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.5 | 3.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.5 | 2.8 | GO:0061198 | fungiform papilla formation(GO:0061198) positive regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000343) positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.3 | 1.4 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.3 | 6.5 | GO:0030213 | hyaluronan biosynthetic process(GO:0030213) |
| 0.1 | 1.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.2 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 4.8 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 1.6 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.5 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 2.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.1 | 6.5 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 4.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.4 | 1.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 3.7 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.2 | 1.5 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.1 | 6.5 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 12.7 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 4.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 1.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.5 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.3 | GO:0048029 | monosaccharide binding(GO:0048029) |
| 0.0 | 1.6 | GO:0001047 | core promoter binding(GO:0001047) |