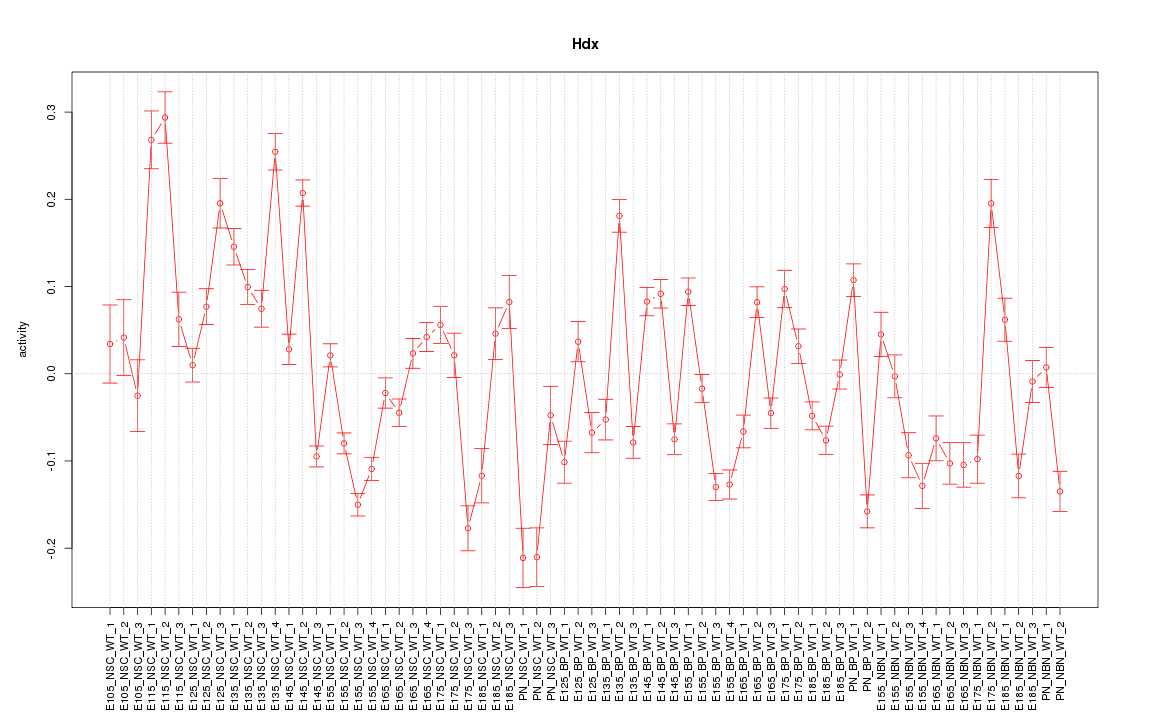
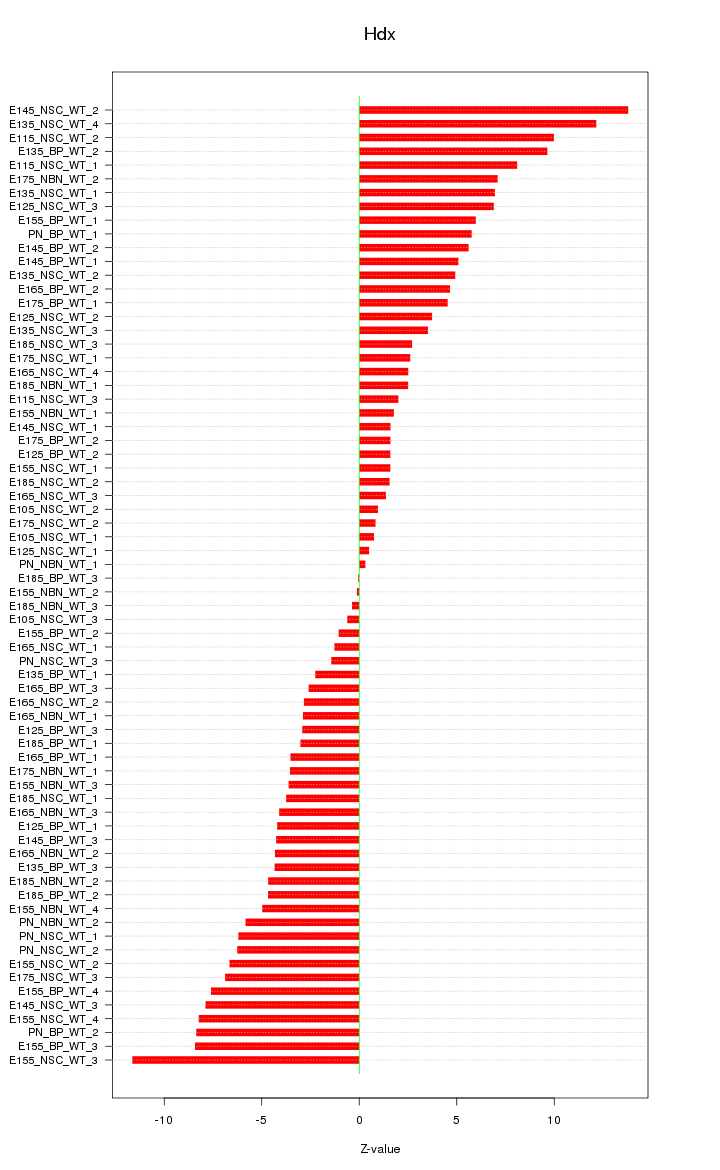
Motif ID: Hdx
Z-value: 5.267
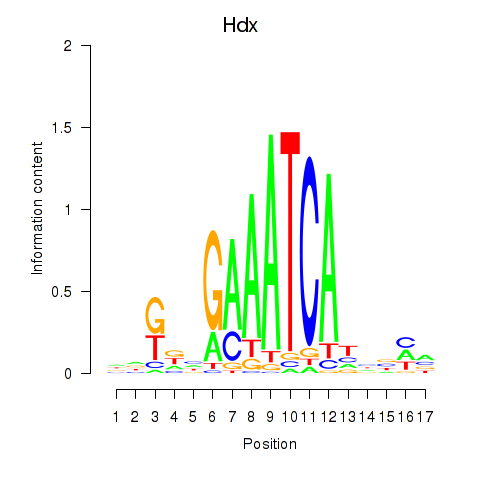
Transcription factors associated with Hdx:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hdx | ENSMUSG00000034551.6 | Hdx |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hdx | mm10_v2_chrX_-_111697069_111697127 | -0.10 | 4.0e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.8 | 52.8 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 4.9 | 14.8 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 4.7 | 14.2 | GO:0046381 | CMP-N-acetylneuraminate metabolic process(GO:0046381) |
| 1.8 | 9.0 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.8 | 7.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 1.6 | 7.9 | GO:0021905 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) |
| 1.2 | 2.4 | GO:1901420 | negative regulation of response to alcohol(GO:1901420) |
| 0.9 | 6.5 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.9 | 4.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) prolactin signaling pathway(GO:0038161) |
| 0.6 | 4.1 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.6 | 2.3 | GO:0006547 | histidine metabolic process(GO:0006547) |
| 0.5 | 1.9 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.5 | 15.0 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.4 | 1.3 | GO:0002295 | T-helper cell lineage commitment(GO:0002295) |
| 0.4 | 21.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.3 | 5.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.2 | 1.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.2 | 4.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.2 | 6.2 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 2.8 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) |
| 0.2 | 3.0 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.1 | 2.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 | 0.4 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 1.1 | GO:0000963 | mitochondrial RNA processing(GO:0000963) |
| 0.1 | 1980.6 | GO:0008150 | biological_process(GO:0008150) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 21.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 1.2 | 4.7 | GO:0032021 | NELF complex(GO:0032021) |
| 1.0 | 5.9 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.9 | 14.8 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.6 | 9.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.4 | 1.3 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.4 | 4.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.3 | 6.5 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 1.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 11.0 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.2 | 2.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.2 | 2.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.2 | 1.9 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 1.0 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 2067.0 | GO:0005575 | cellular_component(GO:0005575) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 14.8 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 4.7 | 14.2 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 2.4 | 7.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 2.3 | 9.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.7 | 5.9 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.7 | 7.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 1.9 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.6 | 11.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.5 | 9.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.5 | 4.6 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.3 | 2.7 | GO:0008193 | tRNA guanylyltransferase activity(GO:0008193) |
| 0.3 | 1.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 2.8 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.2 | 36.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.2 | 1.0 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.2 | 1.3 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.9 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 4.5 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.1 | 1.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2038.4 | GO:0003674 | molecular_function(GO:0003674) |