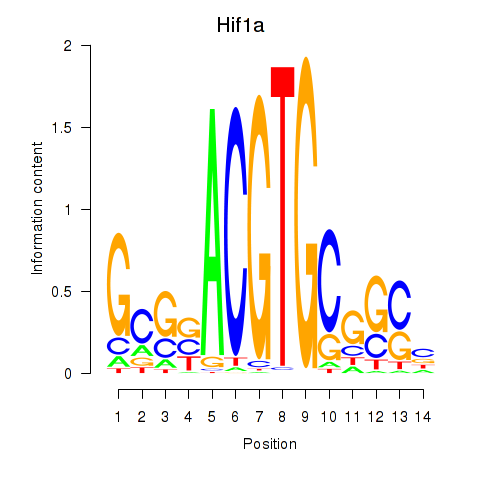
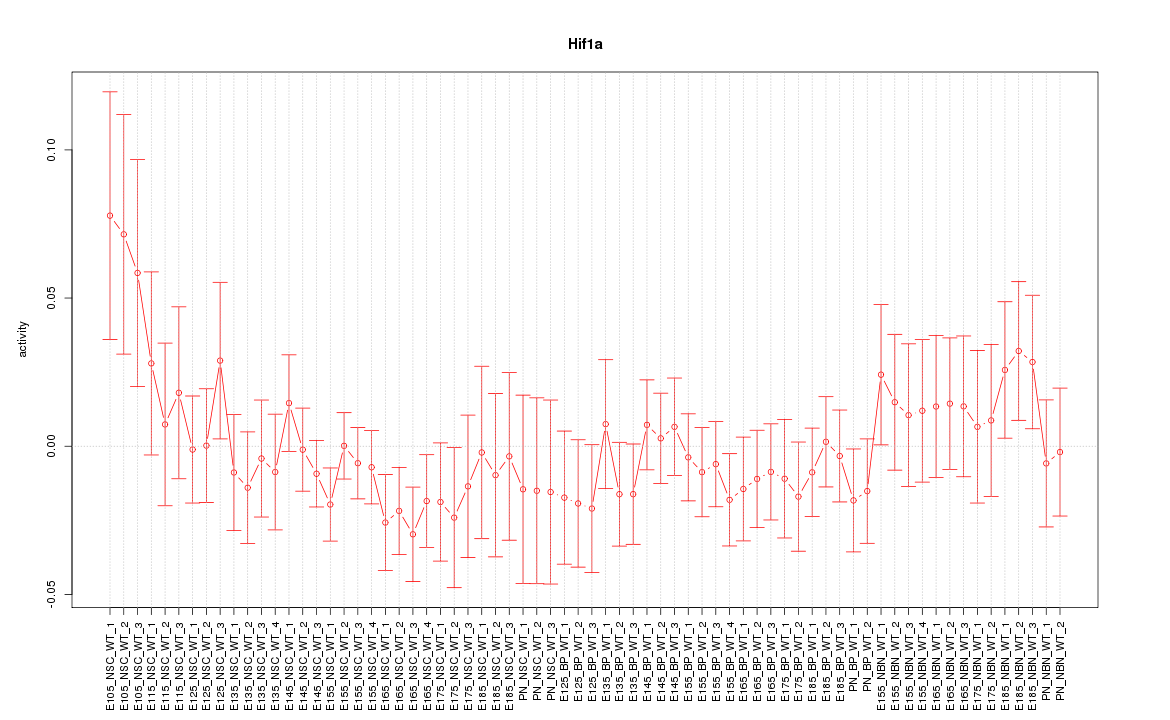
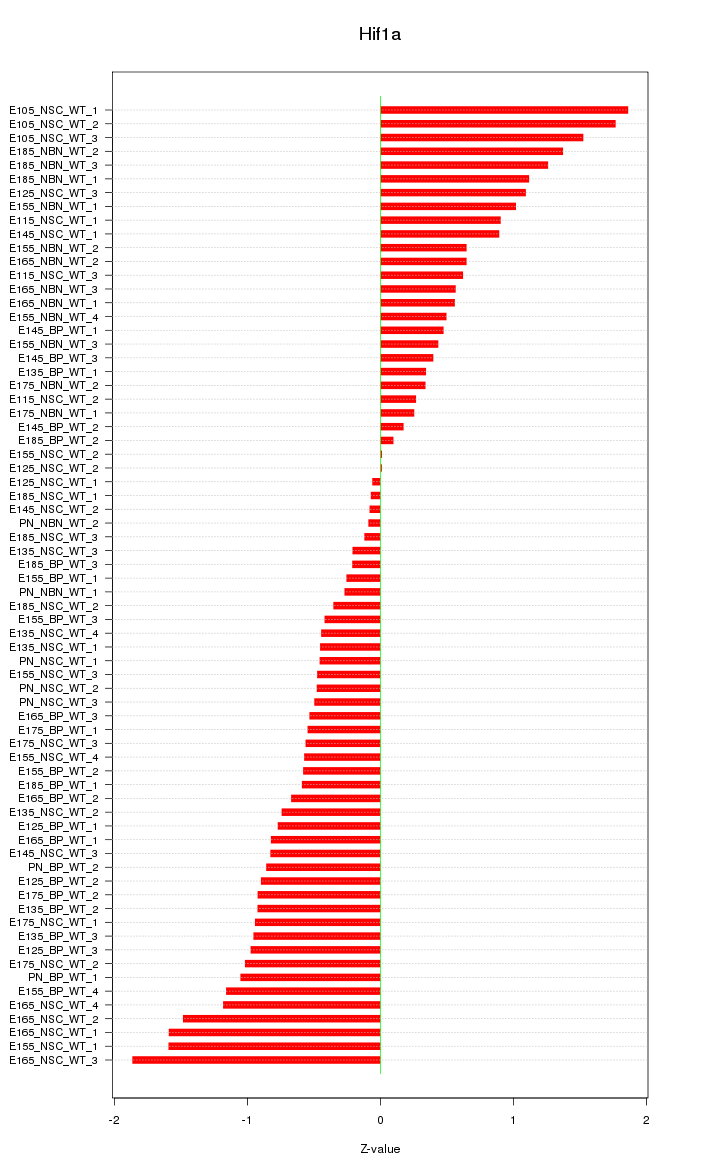
| 2.6 |
7.7 |
GO:1904274 |
tricellular tight junction assembly(GO:1904274) |
| 1.8 |
9.2 |
GO:0019659 |
lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 1.8 |
7.1 |
GO:0010510 |
regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.7 |
1.7 |
GO:0033122 |
regulation of purine nucleotide catabolic process(GO:0033121) negative regulation of purine nucleotide catabolic process(GO:0033122) |
| 1.6 |
6.2 |
GO:0035606 |
peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 1.5 |
4.6 |
GO:0033634 |
positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.5 |
6.0 |
GO:0032439 |
endosome localization(GO:0032439) |
| 1.4 |
4.1 |
GO:0070172 |
positive regulation of tooth mineralization(GO:0070172) |
| 1.2 |
3.7 |
GO:0045819 |
positive regulation of glycogen catabolic process(GO:0045819) |
| 1.0 |
3.1 |
GO:1903225 |
negative regulation of endodermal cell differentiation(GO:1903225) |
| 1.0 |
3.1 |
GO:0014738 |
regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 1.0 |
2.9 |
GO:0009153 |
purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.9 |
1.8 |
GO:0002842 |
T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.8 |
2.4 |
GO:0071336 |
lung growth(GO:0060437) positive regulation of fat cell proliferation(GO:0070346) regulation of hair follicle cell proliferation(GO:0071336) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.6 |
4.2 |
GO:2000124 |
regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.6 |
2.4 |
GO:2000313 |
fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.5 |
4.3 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.5 |
1.9 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.5 |
2.3 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.5 |
1.4 |
GO:0097278 |
complement-dependent cytotoxicity(GO:0097278) |
| 0.5 |
1.4 |
GO:0090071 |
negative regulation of ribosome biogenesis(GO:0090071) |
| 0.4 |
2.2 |
GO:0042117 |
hyaluronan catabolic process(GO:0030214) monocyte activation(GO:0042117) |
| 0.4 |
1.3 |
GO:0008626 |
granzyme-mediated apoptotic signaling pathway(GO:0008626) response to cobalt ion(GO:0032025) |
| 0.4 |
1.2 |
GO:2000224 |
testosterone biosynthetic process(GO:0061370) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.4 |
1.6 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.4 |
2.4 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.4 |
2.7 |
GO:0018401 |
peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.4 |
1.1 |
GO:0006546 |
glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.4 |
3.6 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.4 |
3.6 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.4 |
3.6 |
GO:0030388 |
fructose 6-phosphate metabolic process(GO:0006002) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.3 |
1.7 |
GO:1902570 |
protein localization to nucleolus(GO:1902570) |
| 0.3 |
1.0 |
GO:0070889 |
platelet alpha granule organization(GO:0070889) |
| 0.3 |
2.5 |
GO:0032020 |
ISG15-protein conjugation(GO:0032020) |
| 0.3 |
1.3 |
GO:2000623 |
regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 |
0.5 |
GO:1903519 |
apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.2 |
1.5 |
GO:0035356 |
cellular triglyceride homeostasis(GO:0035356) |
| 0.2 |
1.0 |
GO:0045903 |
positive regulation of translational fidelity(GO:0045903) |
| 0.2 |
1.9 |
GO:0045541 |
negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.2 |
1.7 |
GO:0035948 |
positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) regulation of cellular ketone metabolic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072366) |
| 0.2 |
0.7 |
GO:0007621 |
negative regulation of female receptivity(GO:0007621) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of cellular pH reduction(GO:0032847) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.2 |
2.3 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.2 |
1.7 |
GO:0009435 |
NAD biosynthetic process(GO:0009435) |
| 0.2 |
1.2 |
GO:0045650 |
negative regulation of macrophage differentiation(GO:0045650) |
| 0.2 |
3.4 |
GO:1904816 |
positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.2 |
1.8 |
GO:0042487 |
regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.2 |
0.9 |
GO:0070345 |
negative regulation of fat cell proliferation(GO:0070345) |
| 0.2 |
0.7 |
GO:0019509 |
L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 |
3.6 |
GO:0014002 |
astrocyte development(GO:0014002) |
| 0.2 |
2.1 |
GO:0051085 |
chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.2 |
0.5 |
GO:0033615 |
mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.2 |
0.9 |
GO:0000055 |
ribosomal large subunit export from nucleus(GO:0000055) endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 |
0.5 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
8.0 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 |
4.3 |
GO:0009299 |
mRNA transcription(GO:0009299) |
| 0.1 |
0.7 |
GO:0034214 |
protein hexamerization(GO:0034214) |
| 0.1 |
2.2 |
GO:0048240 |
sperm capacitation(GO:0048240) |
| 0.1 |
0.4 |
GO:1904154 |
positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 |
1.8 |
GO:0000183 |
chromatin silencing at rDNA(GO:0000183) |
| 0.1 |
0.2 |
GO:0021506 |
anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.1 |
5.1 |
GO:0030490 |
maturation of SSU-rRNA(GO:0030490) |
| 0.1 |
0.6 |
GO:1902534 |
single-organism membrane invagination(GO:1902534) |
| 0.1 |
0.4 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.1 |
1.1 |
GO:0042989 |
sequestering of actin monomers(GO:0042989) |
| 0.1 |
0.4 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.1 |
0.3 |
GO:0031590 |
wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 |
3.5 |
GO:0060441 |
epithelial tube branching involved in lung morphogenesis(GO:0060441) branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 |
2.3 |
GO:0071353 |
cellular response to interleukin-4(GO:0071353) |
| 0.1 |
0.4 |
GO:0035553 |
oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.1 |
0.3 |
GO:0042276 |
error-prone translesion synthesis(GO:0042276) |
| 0.1 |
2.4 |
GO:0001706 |
endoderm formation(GO:0001706) |
| 0.1 |
0.3 |
GO:0009223 |
pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.1 |
0.6 |
GO:0006498 |
N-terminal protein lipidation(GO:0006498) |
| 0.1 |
1.1 |
GO:0006670 |
sphingosine metabolic process(GO:0006670) |
| 0.1 |
0.1 |
GO:0061743 |
motor learning(GO:0061743) |
| 0.1 |
2.0 |
GO:0000027 |
ribosomal large subunit assembly(GO:0000027) |
| 0.1 |
4.3 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.1 |
0.3 |
GO:0039536 |
negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.1 |
0.9 |
GO:0034389 |
lipid particle organization(GO:0034389) |
| 0.1 |
6.7 |
GO:0006413 |
translational initiation(GO:0006413) |
| 0.1 |
1.1 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 |
0.8 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.0 |
0.5 |
GO:0010884 |
positive regulation of lipid storage(GO:0010884) |
| 0.0 |
6.7 |
GO:0071229 |
cellular response to acid chemical(GO:0071229) |
| 0.0 |
0.9 |
GO:0099517 |
anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 |
1.5 |
GO:0002181 |
cytoplasmic translation(GO:0002181) |
| 0.0 |
4.8 |
GO:0006364 |
rRNA processing(GO:0006364) |
| 0.0 |
0.8 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.0 |
0.8 |
GO:0042753 |
positive regulation of circadian rhythm(GO:0042753) |
| 0.0 |
0.3 |
GO:0043589 |
skin morphogenesis(GO:0043589) |
| 0.0 |
0.1 |
GO:0070127 |
tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.0 |
1.1 |
GO:0045471 |
response to ethanol(GO:0045471) |
| 0.0 |
0.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.3 |
GO:0000244 |
spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 |
1.5 |
GO:0007601 |
visual perception(GO:0007601) |
| 0.0 |
0.3 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
0.4 |
GO:0043949 |
regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 |
0.2 |
GO:0008272 |
sulfate transport(GO:0008272) |
| 0.0 |
0.6 |
GO:0006506 |
GPI anchor biosynthetic process(GO:0006506) |
| 0.0 |
0.2 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) astral microtubule organization(GO:0030953) |
| 0.0 |
0.6 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |