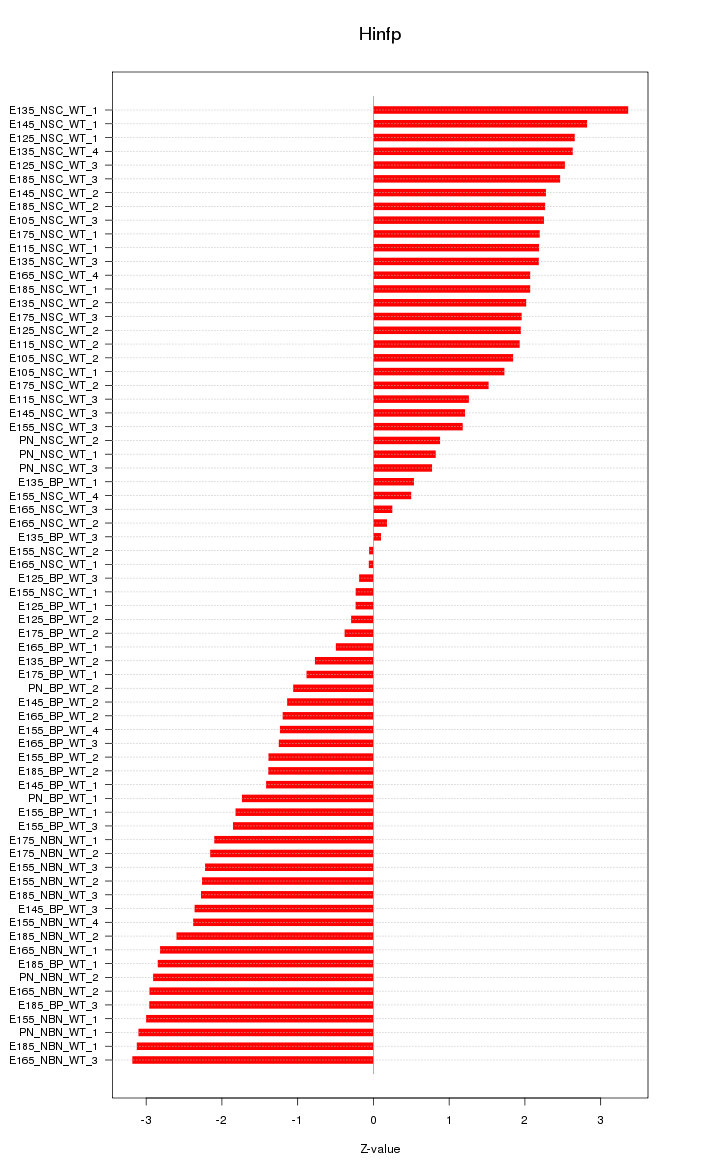
Motif ID: Hinfp
Z-value: 1.937
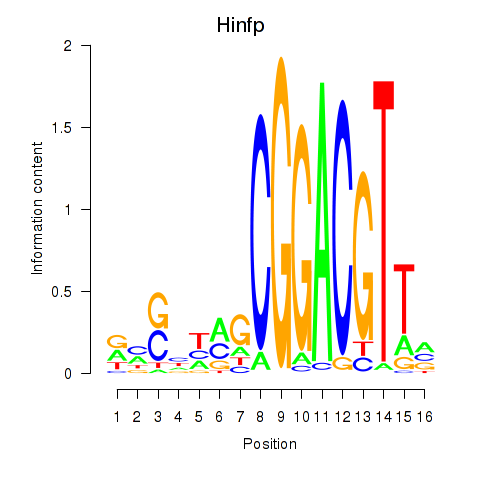
Transcription factors associated with Hinfp:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hinfp | ENSMUSG00000032119.4 | Hinfp |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hinfp | mm10_v2_chr9_-_44305595_44305688 | -0.25 | 3.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 31.6 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 5.5 | 22.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 4.7 | 14.2 | GO:0016321 | female meiosis chromosome segregation(GO:0016321) |
| 4.5 | 22.5 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 4.5 | 13.5 | GO:0030421 | defecation(GO:0030421) |
| 4.4 | 13.2 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 3.9 | 11.8 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 3.6 | 10.8 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 3.3 | 20.0 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 3.3 | 13.2 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 3.1 | 22.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 2.8 | 11.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 2.7 | 8.1 | GO:0060853 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel endothelial cell fate specification(GO:0097101) |
| 2.6 | 7.7 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 2.5 | 7.4 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 2.4 | 12.0 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 2.4 | 7.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 2.3 | 6.8 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 2.2 | 10.9 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 2.2 | 6.5 | GO:0019343 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) |
| 2.0 | 21.5 | GO:0060539 | diaphragm development(GO:0060539) |
| 1.9 | 5.7 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 1.8 | 10.8 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 1.8 | 7.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 1.7 | 7.0 | GO:1903800 | prolactin secretion(GO:0070459) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 1.7 | 6.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.5 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 1.5 | 4.6 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 1.5 | 24.0 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 1.4 | 8.4 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 1.4 | 8.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 1.3 | 20.1 | GO:0009191 | ribonucleoside diphosphate catabolic process(GO:0009191) |
| 1.3 | 9.3 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 1.3 | 3.8 | GO:0048352 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) |
| 1.3 | 11.4 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 1.3 | 5.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 1.2 | 29.9 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 1.2 | 6.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 1.2 | 3.7 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 1.2 | 7.3 | GO:1903755 | regulation of SUMO transferase activity(GO:1903182) positive regulation of SUMO transferase activity(GO:1903755) |
| 1.2 | 6.0 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 1.1 | 13.4 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.1 | 3.3 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 1.1 | 10.9 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 1.0 | 3.0 | GO:0045659 | eosinophil differentiation(GO:0030222) regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 1.0 | 3.0 | GO:0002554 | serotonin secretion by platelet(GO:0002554) interleukin-3 production(GO:0032632) beta selection(GO:0043366) |
| 1.0 | 3.9 | GO:0006776 | vitamin A metabolic process(GO:0006776) positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.9 | 4.7 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.9 | 3.7 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.9 | 4.6 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.9 | 2.7 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.9 | 4.5 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.9 | 3.6 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.9 | 9.6 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.9 | 2.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.9 | 6.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.8 | 6.5 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.8 | 2.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.8 | 6.9 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.7 | 4.9 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.7 | 2.8 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.7 | 8.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 4.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.7 | 6.0 | GO:0006071 | glycerol metabolic process(GO:0006071) |
| 0.6 | 5.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.6 | 3.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 5.0 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.5 | 6.7 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.5 | 7.9 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.5 | 8.6 | GO:0014823 | response to activity(GO:0014823) |
| 0.4 | 2.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.4 | 2.0 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.4 | 7.6 | GO:1904872 | regulation of telomerase RNA localization to Cajal body(GO:1904872) positive regulation of telomerase RNA localization to Cajal body(GO:1904874) |
| 0.4 | 28.9 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.4 | 3.5 | GO:0070475 | nucleolus organization(GO:0007000) rRNA base methylation(GO:0070475) |
| 0.4 | 1.1 | GO:2000574 | regulation of microtubule motor activity(GO:2000574) |
| 0.4 | 6.0 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.4 | 2.3 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.4 | 1.5 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 6.2 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.4 | 2.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.4 | 7.9 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.4 | 5.7 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.4 | 2.8 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.4 | 4.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.3 | 22.7 | GO:0007127 | meiosis I(GO:0007127) |
| 0.3 | 4.8 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.3 | 9.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.3 | 1.7 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.3 | 0.6 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.3 | 3.8 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.3 | 10.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.3 | 9.1 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) DNA replication-independent nucleosome organization(GO:0034724) |
| 0.3 | 4.8 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 1.1 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.3 | 0.5 | GO:0034382 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) |
| 0.3 | 4.9 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.3 | 2.8 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.3 | 12.0 | GO:0070303 | negative regulation of stress-activated MAPK cascade(GO:0032873) negative regulation of stress-activated protein kinase signaling cascade(GO:0070303) |
| 0.2 | 1.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.2 | 2.0 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.2 | 2.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.2 | 0.7 | GO:0052428 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.2 | 1.2 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.2 | 2.8 | GO:0042407 | cristae formation(GO:0042407) |
| 0.2 | 37.7 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.2 | 14.8 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.2 | 9.8 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.2 | 1.9 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.2 | 9.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.2 | 8.0 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 1.5 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.2 | 3.0 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.2 | 2.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.2 | 0.7 | GO:0036481 | intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036481) regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 | 4.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 5.1 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.2 | 6.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 3.0 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 1.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.2 | 1.9 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.2 | 1.5 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 4.4 | GO:0006760 | folic acid-containing compound metabolic process(GO:0006760) |
| 0.1 | 2.4 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 2.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 | 2.5 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.1 | 1.2 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 3.9 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.1 | 7.8 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 12.3 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.1 | 0.3 | GO:0042590 | antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.1 | 4.2 | GO:0006302 | double-strand break repair(GO:0006302) |
| 0.1 | 2.6 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.1 | 2.3 | GO:0001709 | cell fate determination(GO:0001709) |
| 0.1 | 0.4 | GO:0033136 | serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 | 2.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 3.4 | GO:0035315 | hair cell differentiation(GO:0035315) auditory receptor cell differentiation(GO:0042491) |
| 0.1 | 0.6 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.1 | 0.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.1 | 2.6 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 0.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 1.3 | GO:0060008 | Sertoli cell differentiation(GO:0060008) regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.1 | 0.9 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.3 | GO:1903020 | positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.1 | 0.5 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.1 | 2.4 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.9 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.5 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.1 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 1.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 2.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 1.2 | GO:0051193 | regulation of cofactor metabolic process(GO:0051193) |
| 0.0 | 7.9 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 1.1 | GO:0043687 | post-translational protein modification(GO:0043687) |
| 0.0 | 0.5 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 1.3 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 3.1 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 1.1 | GO:0048599 | oocyte development(GO:0048599) |
| 0.0 | 0.7 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.5 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.1 | GO:0090043 | tubulin deacetylation(GO:0090042) regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.4 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.5 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.4 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.5 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 2.0 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 1.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.8 | GO:0006401 | RNA catabolic process(GO:0006401) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.7 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.8 | GO:0000725 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.6 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.3 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 2.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.8 | 20.3 | GO:0036125 | mitochondrial fatty acid beta-oxidation multienzyme complex(GO:0016507) fatty acid beta-oxidation multienzyme complex(GO:0036125) |
| 4.5 | 31.6 | GO:0000796 | condensin complex(GO:0000796) |
| 3.6 | 14.2 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 2.6 | 7.7 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 2.4 | 7.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 2.3 | 7.0 | GO:0031983 | vesicle lumen(GO:0031983) |
| 2.2 | 10.9 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 1.9 | 13.4 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.9 | 5.7 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 1.7 | 10.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.5 | 6.0 | GO:0090537 | CERF complex(GO:0090537) |
| 1.4 | 5.7 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 1.0 | 6.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 1.0 | 6.7 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.9 | 8.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.9 | 12.0 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.8 | 5.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.7 | 3.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.6 | 3.7 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.6 | 8.9 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.6 | 8.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.5 | 2.6 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 1.6 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.5 | 3.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 3.6 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 33.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 10.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.5 | 3.4 | GO:0030677 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.5 | 3.8 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.3 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.4 | 6.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 2.2 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.4 | 6.0 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.4 | 15.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.4 | 31.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.4 | 1.6 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.4 | 1.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.4 | 6.5 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.4 | 2.6 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.4 | 7.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.4 | 3.2 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 2.8 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.3 | 2.3 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.3 | 2.8 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.3 | 8.0 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.3 | 21.3 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.3 | 25.2 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.3 | 4.5 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.3 | 14.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.3 | 16.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 5.6 | GO:0005605 | basal lamina(GO:0005605) |
| 0.3 | 2.9 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 7.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 2.6 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.3 | 3.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 2.0 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.2 | 3.7 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 4.6 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.2 | 7.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.2 | 1.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.2 | 3.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.2 | 11.0 | GO:0005844 | polysome(GO:0005844) |
| 0.2 | 1.5 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.1 | 10.8 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 2.8 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.1 | 5.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 17.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 9.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 5.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.6 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 5.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 3.8 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.5 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 4.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 2.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.1 | 0.6 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.1 | 3.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.8 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 1.9 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.1 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0043205 | fibril(GO:0043205) |
| 0.1 | 4.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.7 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 1.1 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 4.5 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 3.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 4.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.9 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 2.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 4.7 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 2.2 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 7.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 5.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 3.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 4.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 14.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 4.3 | GO:0000790 | nuclear chromatin(GO:0000790) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.4 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 6.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 2.0 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 0.7 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.4 | 21.8 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 4.2 | 37.6 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 4.1 | 16.5 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 3.0 | 12.0 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 2.6 | 7.7 | GO:0034190 | very-low-density lipoprotein particle binding(GO:0034189) apolipoprotein receptor binding(GO:0034190) |
| 2.4 | 21.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 2.4 | 7.2 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 2.4 | 14.2 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.3 | 7.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.2 | 15.2 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 2.2 | 6.5 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 2.1 | 8.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 2.1 | 12.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 2.0 | 9.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 1.8 | 14.5 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 1.7 | 6.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 1.4 | 6.8 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 1.2 | 7.3 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 1.2 | 6.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 1.2 | 6.0 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 1.2 | 9.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 1.1 | 5.6 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 1.0 | 5.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 1.0 | 7.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 1.0 | 3.9 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.9 | 22.7 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.9 | 3.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) serotonin receptor activity(GO:0099589) |
| 0.9 | 8.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.9 | 26.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.8 | 7.4 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.8 | 6.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.8 | 2.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.7 | 8.1 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.7 | 2.8 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.6 | 3.8 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.6 | 3.2 | GO:0043842 | Kdo transferase activity(GO:0043842) |
| 0.6 | 4.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.6 | 2.4 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.5 | 26.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.5 | 8.6 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.5 | 7.9 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.5 | 10.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.5 | 4.6 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.4 | 3.0 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.4 | 2.5 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.4 | 3.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.4 | 2.6 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.4 | 7.1 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.4 | 2.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.4 | 2.6 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 4.0 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.4 | 3.6 | GO:0016920 | pyroglutamyl-peptidase activity(GO:0016920) |
| 0.3 | 22.1 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.3 | 4.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.3 | 3.4 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.3 | 4.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.3 | 8.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.3 | 4.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.3 | 4.9 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.3 | 3.7 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.3 | 5.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 7.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.3 | 13.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.3 | 5.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 11.8 | GO:0061650 | ubiquitin-like protein conjugating enzyme activity(GO:0061650) |
| 0.3 | 0.5 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.2 | 16.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 14.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.9 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.2 | 3.0 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.2 | 4.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.2 | 1.8 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.2 | 2.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.2 | 16.0 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 8.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.2 | 0.5 | GO:0050827 | toxin receptor binding(GO:0050827) |
| 0.2 | 4.2 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.2 | 1.5 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.2 | 9.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 12.7 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.2 | 11.0 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 12.2 | GO:0043851 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (uridine-2'-O-)-methyltransferase activity(GO:0008650) rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) selenocysteine methyltransferase activity(GO:0016205) rRNA (adenine) methyltransferase activity(GO:0016433) rRNA (cytosine) methyltransferase activity(GO:0016434) rRNA (guanine) methyltransferase activity(GO:0016435) 1-phenanthrol methyltransferase activity(GO:0018707) protein-arginine N5-methyltransferase activity(GO:0019702) dimethylarsinite methyltransferase activity(GO:0034541) 4,5-dihydroxybenzo(a)pyrene methyltransferase activity(GO:0034807) 1-hydroxypyrene methyltransferase activity(GO:0034931) 1-hydroxy-6-methoxypyrene methyltransferase activity(GO:0034933) demethylmenaquinone methyltransferase activity(GO:0043770) cobalt-precorrin-6B C5-methyltransferase activity(GO:0043776) cobalt-precorrin-7 C15-methyltransferase activity(GO:0043777) cobalt-precorrin-5B C1-methyltransferase activity(GO:0043780) cobalt-precorrin-3 C17-methyltransferase activity(GO:0043782) dimethylamine methyltransferase activity(GO:0043791) hydroxyneurosporene-O-methyltransferase activity(GO:0043803) tRNA (adenine-57, 58-N(1)-) methyltransferase activity(GO:0043827) methylamine-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043833) trimethylamine methyltransferase activity(GO:0043834) methanol-specific methylcobalamin:coenzyme M methyltransferase activity(GO:0043851) monomethylamine methyltransferase activity(GO:0043852) P-methyltransferase activity(GO:0051994) Se-methyltransferase activity(GO:0051995) 2-phytyl-1,4-naphthoquinone methyltransferase activity(GO:0052624) tRNA (uracil-2'-O-)-methyltransferase activity(GO:0052665) tRNA (cytosine-2'-O-)-methyltransferase activity(GO:0052666) phosphomethylethanolamine N-methyltransferase activity(GO:0052667) tRNA (cytosine-3-)-methyltransferase activity(GO:0052735) rRNA (cytosine-2'-O-)-methyltransferase activity(GO:0070677) rRNA (cytosine-N4-)-methyltransferase activity(GO:0071424) trihydroxyferuloyl spermidine O-methyltransferase activity(GO:0080012) |
| 0.1 | 14.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 13.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.1 | 5.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 1.5 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 3.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 1.5 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.1 | 0.7 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.1 | 1.9 | GO:0034946 | 2-oxoglutaryl-CoA thioesterase activity(GO:0034843) 2,4,4-trimethyl-3-oxopentanoyl-CoA thioesterase activity(GO:0034869) 3-isopropylbut-3-enoyl-CoA thioesterase activity(GO:0034946) glutaryl-CoA hydrolase activity(GO:0044466) |
| 0.1 | 6.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.1 | 3.0 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 7.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 3.9 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.1 | 4.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.1 | 0.5 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 26.0 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.1 | 2.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.1 | 0.8 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.9 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.1 | 0.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.1 | 2.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 27.9 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.1 | 2.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 11.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.6 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 1.7 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 3.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 4.7 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 17.6 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 1.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 2.2 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.0 | 3.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 1.0 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |