Motif ID: Hmga2
Z-value: 1.098
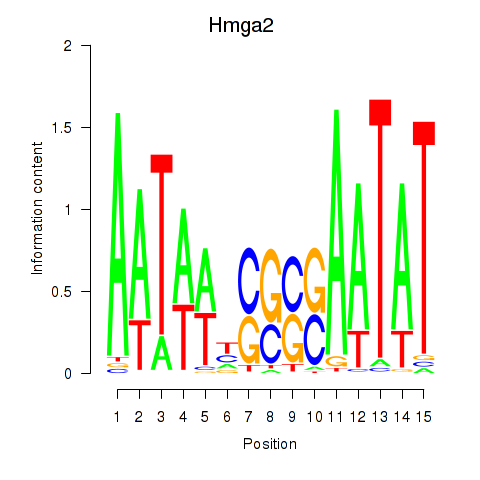
Transcription factors associated with Hmga2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hmga2 | ENSMUSG00000056758.8 | Hmga2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hmga2 | mm10_v2_chr10_-_120476469_120476527 | -0.31 | 1.0e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.8 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.8 | 4.6 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.6 | 1.7 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.5 | 2.6 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.5 | 1.4 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) positive regulation of cortisol secretion(GO:0051464) |
| 0.3 | 1.3 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.3 | 3.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 1.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.2 | 2.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 2.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.2 | 2.3 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 0.6 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.2 | 2.6 | GO:0035635 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.2 | 0.8 | GO:2000427 | regulation of apoptotic cell clearance(GO:2000425) positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.2 | 0.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.2 | 1.3 | GO:2000598 | regulation of cyclin catabolic process(GO:2000598) negative regulation of cyclin catabolic process(GO:2000599) |
| 0.2 | 1.6 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.2 | 0.9 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 | 0.9 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.2 | 0.2 | GO:0006404 | RNA import into nucleus(GO:0006404) |
| 0.1 | 1.0 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 | 0.7 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.1 | 0.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.1 | 1.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 2.0 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.8 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.5 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.1 | 0.9 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 1.0 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.4 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 1.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.5 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 1.2 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 0.4 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 2.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.1 | 0.8 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 6.7 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.1 | 0.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.5 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 1.3 | GO:1900153 | regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900151) positive regulation of nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:1900153) |
| 0.1 | 0.8 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.1 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.9 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 | 0.5 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.2 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 1.0 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 1.2 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:0006862 | nucleotide transport(GO:0006862) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.4 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.7 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) cGMP catabolic process(GO:0046069) |
| 0.0 | 1.4 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.8 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.3 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) |
| 0.0 | 0.6 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.3 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.3 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 4.6 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.7 | 3.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.3 | 2.7 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.3 | 1.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 0.8 | GO:1990415 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.3 | 0.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.2 | 2.6 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.2 | 1.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 0.8 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.2 | 1.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 2.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.5 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.5 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.1 | 0.5 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 2.2 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 3.4 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 0.2 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 4.0 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 1.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 3.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.2 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.9 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.5 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 1.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.3 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 2.8 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 1.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 2.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 1.7 | GO:0005770 | late endosome(GO:0005770) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.9 | 2.6 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.9 | 3.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.6 | 2.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 4.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.3 | 1.0 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.3 | 1.3 | GO:0042895 | antibiotic transporter activity(GO:0042895) |
| 0.2 | 1.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.8 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 1.8 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.2 | 0.8 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 3.7 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.7 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 2.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.5 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 2.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.2 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.1 | 1.1 | GO:0015215 | nucleotide transmembrane transporter activity(GO:0015215) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.0 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 1.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 1.2 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 2.3 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.0 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 1.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 1.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.5 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 4.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.8 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.7 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0003684 | damaged DNA binding(GO:0003684) |





