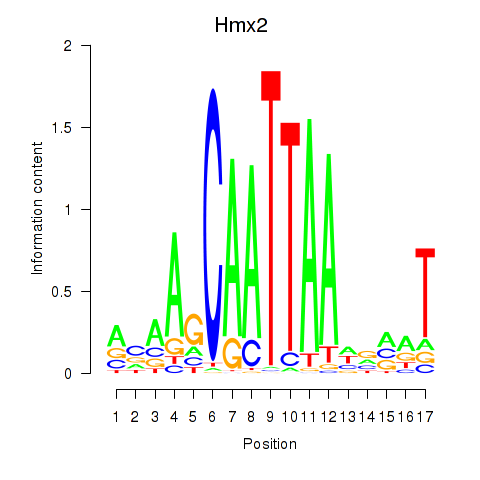
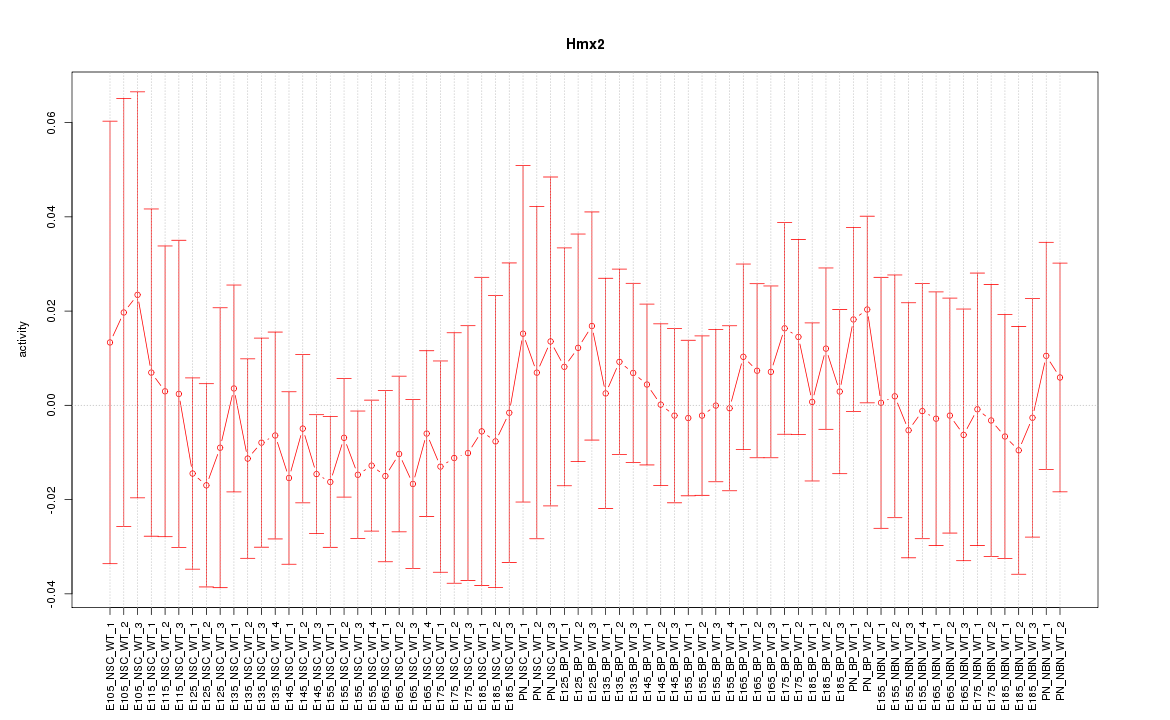
| 0.5 |
1.4 |
GO:0060769 |
positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) kidney smooth muscle tissue development(GO:0072194) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.4 |
1.2 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.4 |
1.5 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.2 |
0.7 |
GO:0035633 |
maintenance of blood-brain barrier(GO:0035633) |
| 0.2 |
0.6 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.2 |
1.1 |
GO:0001887 |
selenium compound metabolic process(GO:0001887) |
| 0.2 |
0.5 |
GO:0015889 |
cobalamin transport(GO:0015889) |
| 0.1 |
0.4 |
GO:0045472 |
response to ether(GO:0045472) |
| 0.1 |
0.4 |
GO:0046544 |
regulation of natural killer cell proliferation(GO:0032817) positive regulation of natural killer cell proliferation(GO:0032819) development of secondary male sexual characteristics(GO:0046544) |
| 0.1 |
0.2 |
GO:0060392 |
negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 |
0.2 |
GO:0035799 |
ureter maturation(GO:0035799) cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.1 |
0.3 |
GO:2001045 |
closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 |
0.3 |
GO:2000680 |
regulation of rubidium ion transport(GO:2000680) |
| 0.1 |
0.7 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 |
0.2 |
GO:2000409 |
positive regulation of T cell extravasation(GO:2000409) |
| 0.1 |
0.3 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.1 |
1.0 |
GO:0006613 |
cotranslational protein targeting to membrane(GO:0006613) |
| 0.1 |
0.2 |
GO:0043651 |
linoleic acid metabolic process(GO:0043651) |
| 0.0 |
0.4 |
GO:0006003 |
fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 |
0.3 |
GO:0006685 |
sphingomyelin catabolic process(GO:0006685) |
| 0.0 |
0.2 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 |
0.3 |
GO:0098734 |
protein depalmitoylation(GO:0002084) positive regulation of pinocytosis(GO:0048549) macromolecule depalmitoylation(GO:0098734) |
| 0.0 |
0.7 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 |
0.2 |
GO:0034080 |
CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 |
0.1 |
GO:0070269 |
pyroptosis(GO:0070269) |
| 0.0 |
0.4 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 |
0.3 |
GO:0046085 |
adenosine metabolic process(GO:0046085) |
| 0.0 |
0.2 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 |
0.1 |
GO:0038128 |
ERBB2 signaling pathway(GO:0038128) |
| 0.0 |
0.7 |
GO:0021854 |
hypothalamus development(GO:0021854) |
| 0.0 |
0.8 |
GO:0015985 |
energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 |
0.2 |
GO:0008611 |
ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.0 |
0.5 |
GO:0006825 |
copper ion transport(GO:0006825) |
| 0.0 |
0.1 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.0 |
0.1 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.0 |
0.1 |
GO:2000348 |
regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 |
0.1 |
GO:0031509 |
telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 |
0.5 |
GO:0043552 |
positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 |
0.2 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 |
0.6 |
GO:0045747 |
positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 |
0.5 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.0 |
0.1 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.2 |
GO:0045945 |
positive regulation of transcription from RNA polymerase III promoter(GO:0045945) negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 |
0.1 |
GO:0048386 |
positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 |
0.1 |
GO:0000028 |
ribosomal small subunit assembly(GO:0000028) |
| 0.0 |
1.0 |
GO:1903955 |
positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 |
0.5 |
GO:0032611 |
interleukin-1 beta production(GO:0032611) |