Motif ID: Hoxa10
Z-value: 0.849
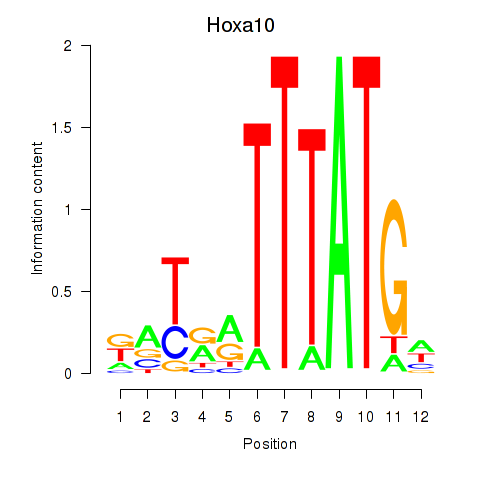
Transcription factors associated with Hoxa10:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxa10 | ENSMUSG00000000938.11 | Hoxa10 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | GO:2000562 | negative regulation of interferon-gamma secretion(GO:1902714) negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 1.6 | 4.8 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.2 | 7.3 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 0.8 | 2.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) terpenoid catabolic process(GO:0016115) primary alcohol catabolic process(GO:0034310) |
| 0.7 | 2.1 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 | 4.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.7 | 4.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.7 | 2.6 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.6 | 5.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.5 | 5.5 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.5 | 1.0 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.5 | 3.2 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 2.3 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.4 | 7.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.4 | 1.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.4 | 1.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.4 | 1.8 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.4 | 2.5 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.3 | 1.6 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.3 | 4.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.3 | 0.8 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.3 | 2.5 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.3 | 3.4 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.3 | 2.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.3 | 0.8 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.3 | 1.3 | GO:0033762 | response to glucagon(GO:0033762) |
| 0.2 | 3.1 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 | 6.6 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.2 | 0.9 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.2 | 0.9 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 2.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.2 | 0.8 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 3.6 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) |
| 0.2 | 4.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.2 | 1.8 | GO:0071493 | cellular response to UV-B(GO:0071493) replicative senescence(GO:0090399) |
| 0.2 | 4.8 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.2 | 0.5 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 4.5 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.1 | 3.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 5.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.8 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 3.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 1.4 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 1.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 | 0.2 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.1 | 1.9 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.1 | 1.1 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.1 | 0.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.7 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 1.1 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.1 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 | 2.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.3 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.1 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.4 | GO:0048741 | skeletal muscle fiber development(GO:0048741) |
| 0.0 | 4.8 | GO:0030010 | establishment of cell polarity(GO:0030010) |
| 0.0 | 0.3 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 1.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.5 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.9 | GO:0015991 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.9 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 1.0 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 1.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 5.9 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.4 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 3.9 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.6 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 1.8 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 1.7 | GO:0010506 | regulation of autophagy(GO:0010506) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.6 | 1.8 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.5 | 3.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.5 | 4.8 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.4 | 2.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.4 | 2.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.3 | 2.3 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.3 | 2.2 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.3 | 5.4 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.5 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 2.6 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.2 | 3.6 | GO:0043196 | varicosity(GO:0043196) |
| 0.2 | 1.3 | GO:0045275 | respiratory chain complex III(GO:0045275) |
| 0.2 | 0.9 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.1 | 1.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 4.3 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 1.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 0.9 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 3.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 0.9 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 5.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 5.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 1.9 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.9 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 2.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 4.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.8 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.8 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.7 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 3.4 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 1.9 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 1.9 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 4.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 7.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 5.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 2.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 1.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 1.3 | 5.1 | GO:0086038 | calcium:sodium antiporter activity involved in regulation of cardiac muscle cell membrane potential(GO:0086038) |
| 1.1 | 7.6 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 1.0 | 8.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.9 | 5.4 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.9 | 1.8 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.7 | 2.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.7 | 4.7 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.6 | 2.2 | GO:0001639 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) |
| 0.5 | 4.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.5 | 1.9 | GO:0004954 | icosanoid receptor activity(GO:0004953) prostanoid receptor activity(GO:0004954) prostaglandin receptor activity(GO:0004955) |
| 0.4 | 2.5 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.4 | 3.2 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 1.1 | GO:0004942 | anaphylatoxin receptor activity(GO:0004942) |
| 0.4 | 2.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.3 | 5.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.3 | 5.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 2.5 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.3 | 0.8 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 4.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 1.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.2 | 0.7 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 2.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 0.9 | GO:0071532 | ornithine decarboxylase inhibitor activity(GO:0008073) ankyrin repeat binding(GO:0071532) |
| 0.2 | 4.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 1.0 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.2 | 2.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 1.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.1 | 0.4 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 1.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 1.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.8 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.1 | 3.4 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 0.8 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 1.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 3.1 | GO:0004120 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) photoreceptor cyclic-nucleotide phosphodiesterase activity(GO:0004120) 7,8-dihydro-D-neopterin 2',3'-cyclic phosphate phosphodiesterase activity(GO:0044688) inositol phosphosphingolipid phospholipase activity(GO:0052712) inositol phosphorylceramide phospholipase activity(GO:0052713) mannosyl-inositol phosphorylceramide phospholipase activity(GO:0052714) mannosyl-diinositol phosphorylceramide phospholipase activity(GO:0052715) |
| 0.1 | 3.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 4.5 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 1.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.1 | 12.7 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.5 | GO:0052872 | 3-(3-hydroxyphenyl)propionate hydroxylase activity(GO:0008688) 4-chlorobenzaldehyde oxidase activity(GO:0018471) 3,5-xylenol methylhydroxylase activity(GO:0018630) phenylacetate hydroxylase activity(GO:0018631) 4-nitrophenol 4-monooxygenase activity(GO:0018632) dimethyl sulfide monooxygenase activity(GO:0018633) alpha-pinene monooxygenase [NADH] activity(GO:0018634) phenanthrene 9,10-monooxygenase activity(GO:0018636) 1-hydroxy-2-naphthoate hydroxylase activity(GO:0018637) toluene 4-monooxygenase activity(GO:0018638) xylene monooxygenase activity(GO:0018639) dibenzothiophene monooxygenase activity(GO:0018640) 6-hydroxy-3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018641) chlorophenol 4-monooxygenase activity(GO:0018642) carbon disulfide oxygenase activity(GO:0018643) toluene 2-monooxygenase activity(GO:0018644) 1-hydroxy-2-oxolimonene 1,2-monooxygenase activity(GO:0018646) phenanthrene 1,2-monooxygenase activity(GO:0018647) tetrahydrofuran hydroxylase activity(GO:0018649) styrene monooxygenase activity(GO:0018650) toluene-4-sulfonate monooxygenase activity(GO:0018651) toluene-sulfonate methyl-monooxygenase activity(GO:0018652) 3-methyl-2-oxo-1,2-dihydroquinoline 6-monooxygenase activity(GO:0018653) 2-hydroxy-phenylacetate hydroxylase activity(GO:0018654) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA 1,2-monooxygenase activity(GO:0018655) phenanthrene 3,4-monooxygenase activity(GO:0018656) toluene 3-monooxygenase activity(GO:0018657) 4-hydroxyphenylacetate,NADH:oxygen oxidoreductase (3-hydroxylating) activity(GO:0018660) limonene monooxygenase activity(GO:0019113) 2-methylnaphthalene hydroxylase activity(GO:0034526) 1-methylnaphthalene hydroxylase activity(GO:0034534) bisphenol A hydroxylase A activity(GO:0034560) salicylate 5-hydroxylase activity(GO:0034785) isobutylamine N-hydroxylase activity(GO:0034791) branched-chain dodecylbenzene sulfonate monooxygenase activity(GO:0034802) 3-HSA hydroxylase activity(GO:0034819) 4-hydroxypyridine-3-hydroxylase activity(GO:0034894) 2-octaprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase activity(GO:0043719) 6-hydroxynicotinate 3-monooxygenase activity(GO:0043731) tocotrienol omega-hydroxylase activity(GO:0052872) thalianol hydroxylase activity(GO:0080014) |
| 0.0 | 3.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 2.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.8 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 1.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.8 | GO:0035380 | C-3 sterol dehydrogenase (C-4 sterol decarboxylase) activity(GO:0000252) mevaldate reductase activity(GO:0004495) gluconate dehydrogenase activity(GO:0008875) epoxide dehydrogenase activity(GO:0018451) 5-exo-hydroxycamphor dehydrogenase activity(GO:0018452) 2-hydroxytetrahydrofuran dehydrogenase activity(GO:0018453) acetoin dehydrogenase activity(GO:0019152) phenylcoumaran benzylic ether reductase activity(GO:0032442) D-xylose:NADP reductase activity(GO:0032866) L-arabinose:NADP reductase activity(GO:0032867) D-arabinitol dehydrogenase, D-ribulose forming (NADP+) activity(GO:0033709) (R)-(-)-1,2,3,4-tetrahydronaphthol dehydrogenase activity(GO:0034831) 3-hydroxymenthone dehydrogenase activity(GO:0034840) very long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0035380) dihydrotestosterone 17-beta-dehydrogenase activity(GO:0035410) (R)-2-hydroxyisocaproate dehydrogenase activity(GO:0043713) L-arabinose 1-dehydrogenase (NADP+) activity(GO:0044103) L-xylulose reductase (NAD+) activity(GO:0044105) 3-ketoglucose-reductase activity(GO:0048258) D-arabinitol dehydrogenase, D-xylulose forming (NADP+) activity(GO:0052677) |
| 0.0 | 3.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 1.2 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 3.3 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 1.6 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 2.1 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 4.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0002039 | p53 binding(GO:0002039) |





