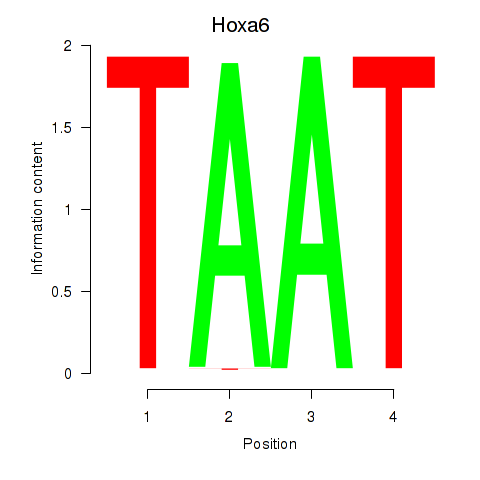
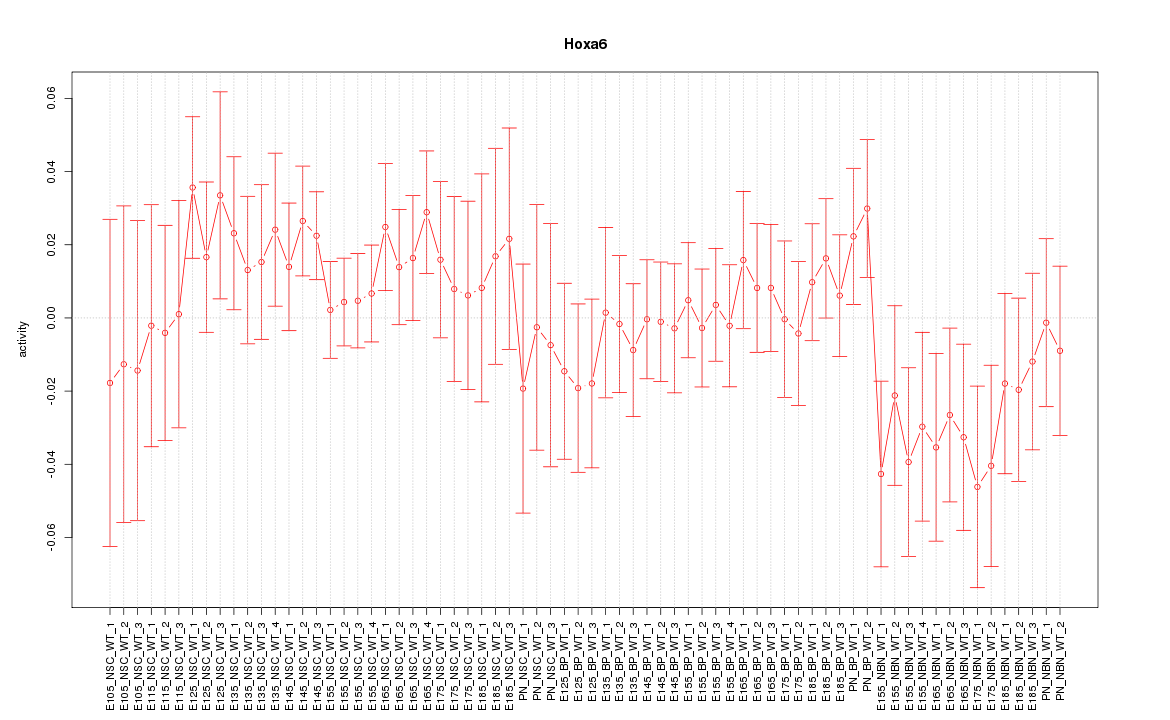
| 7.5 |
22.4 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 3.9 |
11.7 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 1.9 |
11.4 |
GO:0006570 |
tyrosine metabolic process(GO:0006570) |
| 1.7 |
5.0 |
GO:0060023 |
soft palate development(GO:0060023) |
| 1.5 |
6.0 |
GO:0046552 |
eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 1.4 |
11.6 |
GO:0007406 |
negative regulation of neuroblast proliferation(GO:0007406) |
| 1.3 |
3.9 |
GO:0042320 |
regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 1.3 |
9.0 |
GO:0071699 |
olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 1.2 |
4.9 |
GO:0043490 |
malate-aspartate shuttle(GO:0043490) |
| 1.2 |
6.0 |
GO:0007223 |
Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 1.2 |
14.2 |
GO:0060501 |
positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 1.0 |
3.1 |
GO:0060423 |
limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) negative regulation of mitotic cell cycle, embryonic(GO:0045976) foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 1.0 |
8.0 |
GO:0048664 |
neuron fate determination(GO:0048664) |
| 1.0 |
10.6 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.8 |
2.5 |
GO:0003310 |
pancreatic A cell differentiation(GO:0003310) |
| 0.8 |
3.9 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.7 |
6.6 |
GO:0097154 |
GABAergic neuron differentiation(GO:0097154) |
| 0.7 |
11.0 |
GO:0048387 |
negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 |
7.1 |
GO:0060539 |
diaphragm development(GO:0060539) |
| 0.6 |
5.0 |
GO:0045199 |
maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.6 |
4.3 |
GO:0010908 |
regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.5 |
5.3 |
GO:0002118 |
aggressive behavior(GO:0002118) |
| 0.5 |
2.7 |
GO:1903215 |
negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.5 |
16.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.4 |
2.6 |
GO:0032229 |
positive regulation of gamma-aminobutyric acid secretion(GO:0014054) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.4 |
1.3 |
GO:0035582 |
sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.4 |
3.4 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) |
| 0.4 |
2.2 |
GO:0060385 |
axonogenesis involved in innervation(GO:0060385) |
| 0.3 |
2.4 |
GO:0048251 |
elastic fiber assembly(GO:0048251) |
| 0.3 |
7.7 |
GO:0035855 |
megakaryocyte development(GO:0035855) |
| 0.3 |
0.8 |
GO:0046013 |
regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 |
4.9 |
GO:0036342 |
post-anal tail morphogenesis(GO:0036342) |
| 0.2 |
5.6 |
GO:0032967 |
positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 |
0.7 |
GO:0090327 |
negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.2 |
0.8 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.2 |
2.0 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 |
0.6 |
GO:0021562 |
vestibulocochlear nerve development(GO:0021562) |
| 0.2 |
0.2 |
GO:0003357 |
noradrenergic neuron differentiation(GO:0003357) |
| 0.2 |
6.2 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.2 |
3.1 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.2 |
0.8 |
GO:0034196 |
acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.2 |
2.0 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.1 |
1.6 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 |
1.3 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 |
0.3 |
GO:0033484 |
nitric oxide homeostasis(GO:0033484) |
| 0.1 |
0.6 |
GO:0038032 |
termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 |
6.2 |
GO:0006940 |
regulation of smooth muscle contraction(GO:0006940) |
| 0.1 |
9.4 |
GO:0043488 |
regulation of mRNA stability(GO:0043488) |
| 0.1 |
1.8 |
GO:0090036 |
regulation of protein kinase C signaling(GO:0090036) |
| 0.1 |
0.4 |
GO:0032049 |
cardiolipin biosynthetic process(GO:0032049) |
| 0.1 |
0.8 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.1 |
2.1 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
0.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
2.7 |
GO:0030901 |
midbrain development(GO:0030901) |
| 0.1 |
0.5 |
GO:0009249 |
protein lipoylation(GO:0009249) |
| 0.1 |
0.5 |
GO:0036233 |
glycine import(GO:0036233) |
| 0.1 |
0.3 |
GO:2000504 |
positive regulation of blood vessel remodeling(GO:2000504) |
| 0.1 |
0.8 |
GO:0021521 |
ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 |
0.8 |
GO:0021542 |
dentate gyrus development(GO:0021542) |
| 0.1 |
1.1 |
GO:0001833 |
inner cell mass cell proliferation(GO:0001833) |
| 0.1 |
0.5 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 |
0.4 |
GO:0090074 |
negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 |
0.7 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 |
1.8 |
GO:1900087 |
positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 |
0.2 |
GO:0055071 |
cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 |
1.0 |
GO:0007617 |
mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 |
1.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
4.9 |
GO:0006261 |
DNA-dependent DNA replication(GO:0006261) |
| 0.0 |
0.9 |
GO:0010569 |
regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 |
0.1 |
GO:2000253 |
positive regulation of feeding behavior(GO:2000253) |
| 0.0 |
0.9 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
4.2 |
GO:0007605 |
sensory perception of sound(GO:0007605) |
| 0.0 |
1.1 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.6 |
GO:0009954 |
proximal/distal pattern formation(GO:0009954) |
| 0.0 |
2.1 |
GO:0007156 |
homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 |
0.4 |
GO:0033194 |
response to hydroperoxide(GO:0033194) |
| 0.0 |
1.3 |
GO:0022900 |
electron transport chain(GO:0022900) |
| 0.0 |
0.1 |
GO:0006686 |
sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 |
0.2 |
GO:0048665 |
neuron fate specification(GO:0048665) |
| 0.0 |
0.2 |
GO:0061014 |
positive regulation of mRNA catabolic process(GO:0061014) |
| 0.0 |
0.4 |
GO:0060135 |
maternal process involved in female pregnancy(GO:0060135) |