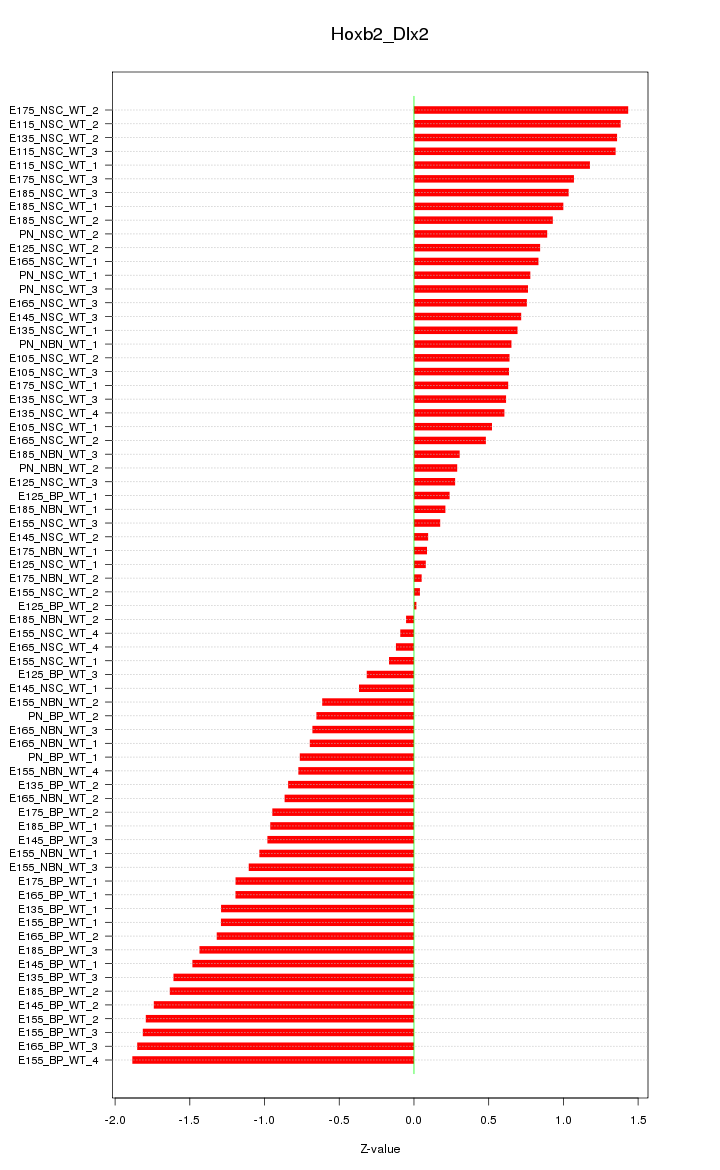
Motif ID: Hoxb2_Dlx2
Z-value: 0.964
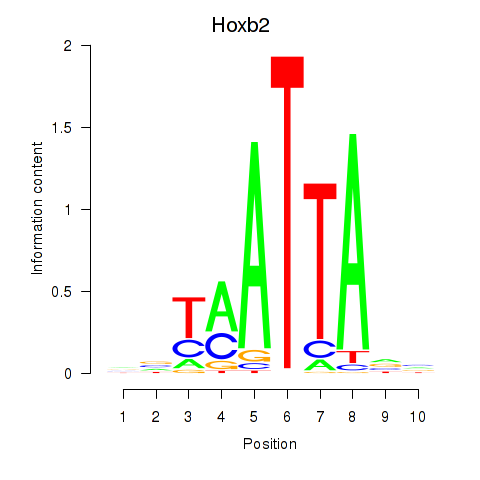
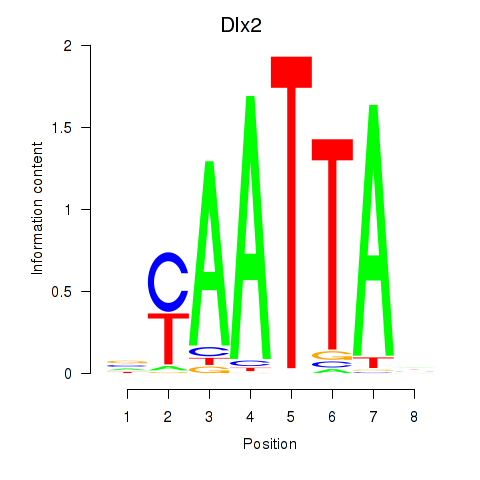
Transcription factors associated with Hoxb2_Dlx2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Dlx2 | ENSMUSG00000023391.7 | Dlx2 |
| Hoxb2 | ENSMUSG00000075588.5 | Hoxb2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx2 | mm10_v2_chr2_-_71546745_71546758 | 0.86 | 3.4e-21 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.9 | 29.7 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 6.0 | 24.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 5.3 | 21.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 3.6 | 10.7 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 2.9 | 11.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 2.8 | 8.4 | GO:0048818 | embryonic nail plate morphogenesis(GO:0035880) positive regulation of hair follicle maturation(GO:0048818) positive regulation of catagen(GO:0051795) frontal suture morphogenesis(GO:0060364) |
| 1.8 | 5.5 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.6 | 4.9 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.6 | 4.9 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 1.4 | 8.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 1.4 | 5.4 | GO:0072199 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 1.3 | 8.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 1.3 | 4.0 | GO:0072204 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) |
| 1.3 | 5.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 1.2 | 9.9 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 1.2 | 3.6 | GO:1905065 | cysteine biosynthetic process via cystathionine(GO:0019343) cysteine biosynthetic process(GO:0019344) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 1.2 | 3.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 1.1 | 3.2 | GO:0008228 | opsonization(GO:0008228) modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 1.0 | 6.8 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.9 | 2.6 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.8 | 5.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.8 | 3.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.8 | 24.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.7 | 3.6 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.7 | 9.2 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.7 | 4.8 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.7 | 9.5 | GO:0007530 | sex determination(GO:0007530) |
| 0.7 | 2.0 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.7 | 2.7 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.7 | 2.6 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.6 | 3.9 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.6 | 7.1 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.6 | 3.0 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.6 | 5.4 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.6 | 5.3 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.6 | 9.4 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.6 | 1.8 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.6 | 3.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.6 | 9.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.5 | 14.1 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.5 | 1.0 | GO:0060023 | soft palate development(GO:0060023) |
| 0.4 | 1.3 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.4 | 2.1 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.4 | 1.2 | GO:0090526 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 1.9 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.4 | 2.6 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.4 | 2.6 | GO:0015791 | polyol transport(GO:0015791) urea transport(GO:0015840) bile acid secretion(GO:0032782) urea transmembrane transport(GO:0071918) |
| 0.4 | 2.9 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.4 | 1.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.4 | 1.8 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.3 | 8.5 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.3 | 5.8 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.3 | 2.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.3 | 0.9 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.3 | 4.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.3 | 0.8 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 | 1.7 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.3 | 3.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.3 | 3.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.3 | 2.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.2 | 0.9 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.2 | 0.2 | GO:0045852 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.2 | 3.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.2 | 2.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 3.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.2 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 5.3 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.2 | 1.4 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.2 | 0.9 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.9 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.1 | 2.9 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 0.2 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.1 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 | 7.6 | GO:0042475 | odontogenesis of dentin-containing tooth(GO:0042475) |
| 0.1 | 13.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 6.3 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 2.1 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.1 | 6.1 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.1 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.1 | 3.6 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.1 | 1.1 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 3.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 9.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.1 | 6.2 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.1 | 1.7 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 2.9 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.1 | 1.3 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 | 1.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 6.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 0.9 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 0.6 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 1.7 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.1 | 0.3 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.7 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.1 | 4.8 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.9 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 2.1 | GO:0019915 | lipid storage(GO:0019915) |
| 0.1 | 3.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 | 2.9 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.1 | 0.2 | GO:1902915 | progesterone receptor signaling pathway(GO:0050847) negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 2.5 | GO:0071774 | response to fibroblast growth factor(GO:0071774) |
| 0.1 | 3.1 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 1.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.0 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 5.4 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.3 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.1 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 1.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 1.0 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 3.9 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.7 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.0 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.3 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.7 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 7.3 | GO:0007059 | chromosome segregation(GO:0007059) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 3.4 | GO:0001501 | skeletal system development(GO:0001501) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.1 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.2 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.2 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.4 | GO:0050868 | negative regulation of T cell activation(GO:0050868) |
| 0.0 | 0.5 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 4.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 2.0 | 11.8 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 1.6 | 4.9 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 1.4 | 24.1 | GO:0043205 | fibril(GO:0043205) |
| 1.4 | 8.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 1.0 | 8.4 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 10.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.5 | 2.6 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 2.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.4 | 2.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 3.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.3 | 7.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.3 | 9.2 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.2 | 13.2 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 1.9 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 1.0 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 3.1 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.2 | 1.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.2 | 4.3 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 1.0 | GO:0061689 | paranodal junction(GO:0033010) tricellular tight junction(GO:0061689) |
| 0.2 | 5.0 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 9.1 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 0.7 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 3.5 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 3.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 5.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.1 | 1.7 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 1.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 1.4 | GO:0032797 | SMN complex(GO:0032797) |
| 0.1 | 0.7 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.1 | 3.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 1.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 1.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 3.0 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 0.2 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.1 | 0.8 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.1 | 22.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 3.6 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.9 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 9.6 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.2 | GO:0051286 | cell tip(GO:0051286) |
| 0.0 | 8.0 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.8 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 1.2 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.4 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.0 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.9 | GO:0000313 | organellar ribosome(GO:0000313) mitochondrial ribosome(GO:0005761) |
| 0.0 | 86.5 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 10.4 | GO:0005615 | extracellular space(GO:0005615) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 24.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.3 | 11.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 1.3 | 10.7 | GO:0050693 | LBD domain binding(GO:0050693) |
| 1.2 | 9.9 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 1.2 | 3.6 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 1.1 | 3.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 1.0 | 10.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 1.0 | 5.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 1.0 | 2.9 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.9 | 11.8 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.8 | 3.0 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.7 | 3.6 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.7 | 5.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.7 | 5.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.6 | 3.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 6.8 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.6 | 8.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.5 | 12.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.5 | 8.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.5 | 2.4 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.5 | 3.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.4 | 2.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.4 | 4.0 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.4 | 2.2 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.4 | 2.6 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.4 | 2.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.4 | 2.9 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.4 | 9.6 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.4 | 1.9 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.4 | 4.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 2.6 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.4 | 3.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.3 | 4.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.3 | 2.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.3 | 7.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 6.2 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.3 | 3.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.3 | 4.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.3 | 13.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.3 | 3.6 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.3 | 8.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.2 | 6.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 2.1 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.2 | 0.9 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 0.6 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.2 | 12.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 1.2 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 0.8 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 5.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 3.5 | GO:0016208 | AMP binding(GO:0016208) |
| 0.2 | 47.1 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 2.9 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 12.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.7 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.9 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.5 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.9 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.1 | 1.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.1 | 1.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 11.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 2.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 13.5 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) |
| 0.1 | 2.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 48.4 | GO:0043565 | sequence-specific DNA binding(GO:0043565) |
| 0.1 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 2.6 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.7 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 2.8 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.7 | GO:0046933 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 1.1 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.7 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.9 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.6 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 8.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 1.9 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0016749 | N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 1.0 | GO:0032934 | cholesterol binding(GO:0015485) sterol binding(GO:0032934) |
| 0.0 | 2.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 3.9 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 1.5 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.5 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 2.0 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 1.2 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.2 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |