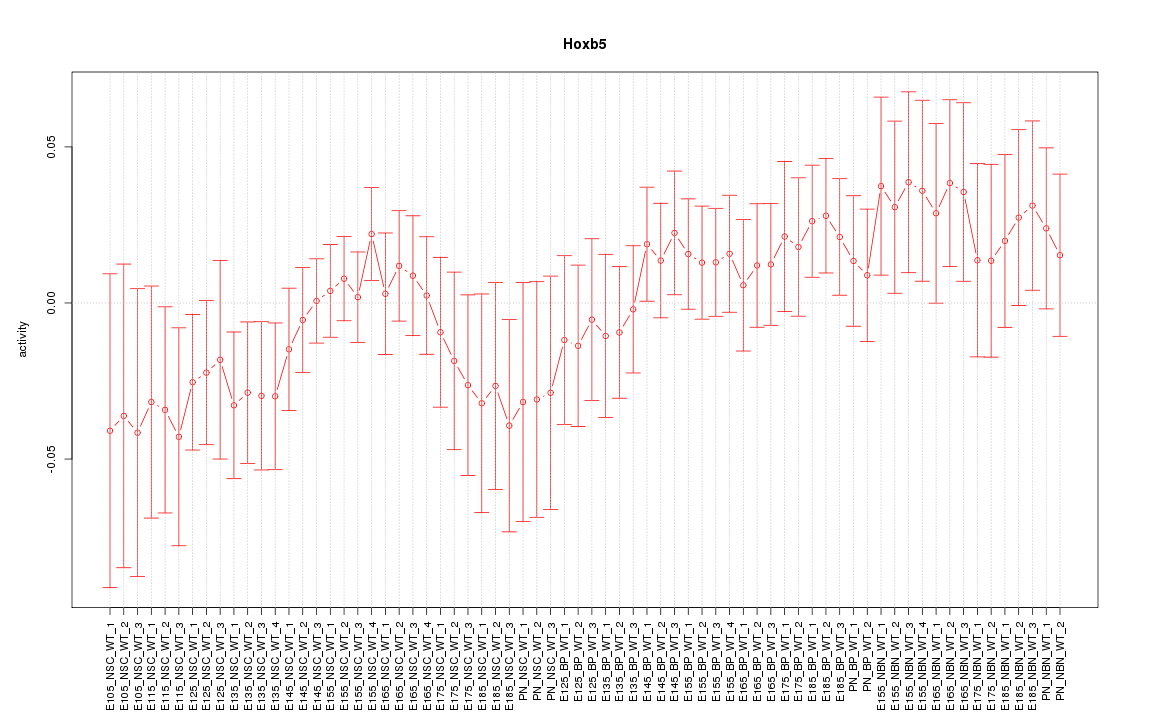
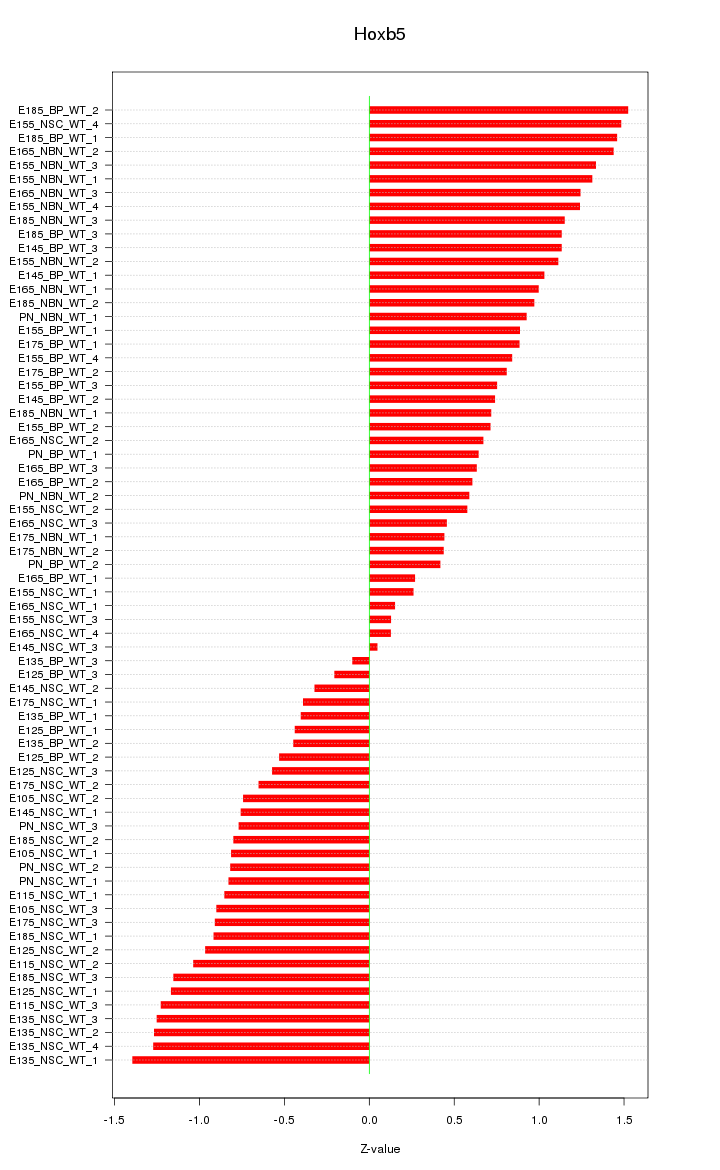
Motif ID: Hoxb5
Z-value: 0.887
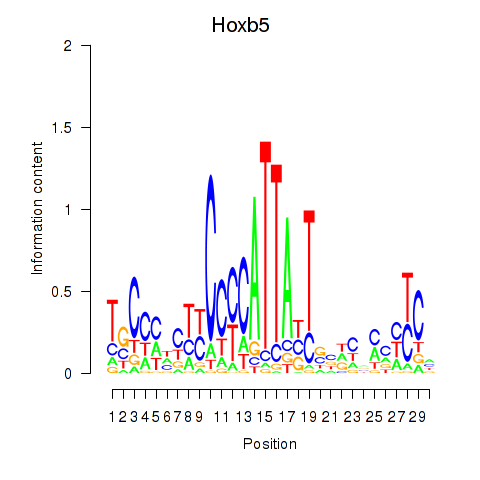
Transcription factors associated with Hoxb5:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxb5 | ENSMUSG00000038700.3 | Hoxb5 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.6 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 1.3 | 9.2 | GO:0071361 | cellular response to zinc ion(GO:0071294) cellular response to ethanol(GO:0071361) |
| 1.3 | 5.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.8 | 2.5 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.7 | 3.6 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.6 | 10.0 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.6 | 1.7 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.6 | 3.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.6 | 1.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.4 | 12.2 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.4 | 4.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 6.4 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.3 | 4.9 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.2 | 4.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.2 | 3.8 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.2 | 1.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.2 | 1.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 1.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 | 0.7 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.2 | 4.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.2 | 2.0 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.1 | 2.4 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.1 | 1.8 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.1 | 15.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.1 | 0.5 | GO:0043651 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) |
| 0.1 | 3.0 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 0.7 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.1 | 3.6 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 0.3 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.1 | 4.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.4 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.0 | 1.0 | GO:0008299 | isoprenoid biosynthetic process(GO:0008299) |
| 0.0 | 1.8 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 3.5 | GO:0019233 | sensory perception of pain(GO:0019233) |
| 0.0 | 1.4 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 3.4 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.3 | GO:0033692 | cellular polysaccharide biosynthetic process(GO:0033692) |
| 0.0 | 1.8 | GO:1903169 | regulation of calcium ion transmembrane transport(GO:1903169) |
| 0.0 | 2.5 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.2 | GO:2001257 | regulation of cation channel activity(GO:2001257) |
| 0.0 | 0.1 | GO:0045583 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.0 | 10.7 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 1.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.7 | GO:0001824 | blastocyst development(GO:0001824) |
| 0.0 | 9.3 | GO:0007017 | microtubule-based process(GO:0007017) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 11.6 | GO:0033269 | internode region of axon(GO:0033269) |
| 1.0 | 4.9 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.5 | 3.6 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.4 | 4.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 3.0 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.2 | 9.4 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 2.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 3.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 4.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 10.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 4.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 4.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 8.3 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 1.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 5.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.7 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 2.5 | GO:0044450 | microtubule organizing center part(GO:0044450) |
| 0.0 | 10.1 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 5.2 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 4.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.1 | 12.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 3.1 | 9.2 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 1.5 | 12.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 1.2 | 6.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.0 | 4.9 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.9 | 5.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.5 | 1.6 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.4 | 1.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.3 | 1.0 | GO:0048045 | 4-hydroxybenzoate octaprenyltransferase activity(GO:0008412) protoheme IX farnesyltransferase activity(GO:0008495) (S)-2,3-di-O-geranylgeranylglyceryl phosphate synthase activity(GO:0043888) cadaverine aminopropyltransferase activity(GO:0043918) agmatine aminopropyltransferase activity(GO:0043919) 1,4-dihydroxy-2-naphthoate octaprenyltransferase activity(GO:0046428) trans-pentaprenyltranstransferase activity(GO:0048045) ATP dimethylallyltransferase activity(GO:0052622) ADP dimethylallyltransferase activity(GO:0052623) |
| 0.3 | 5.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.3 | 4.9 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 1.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.8 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.2 | 2.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 0.7 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.2 | 1.9 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 4.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 3.4 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 3.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.2 | 1.2 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 11.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.4 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.1 | 3.5 | GO:0042165 | neurotransmitter binding(GO:0042165) |
| 0.1 | 15.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 2.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 3.6 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.0 | 2.9 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 1.9 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 4.2 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.0 | 3.0 | GO:0015078 | hydrogen ion transmembrane transporter activity(GO:0015078) |
| 0.0 | 3.4 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 2.6 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 1.4 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 2.4 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |