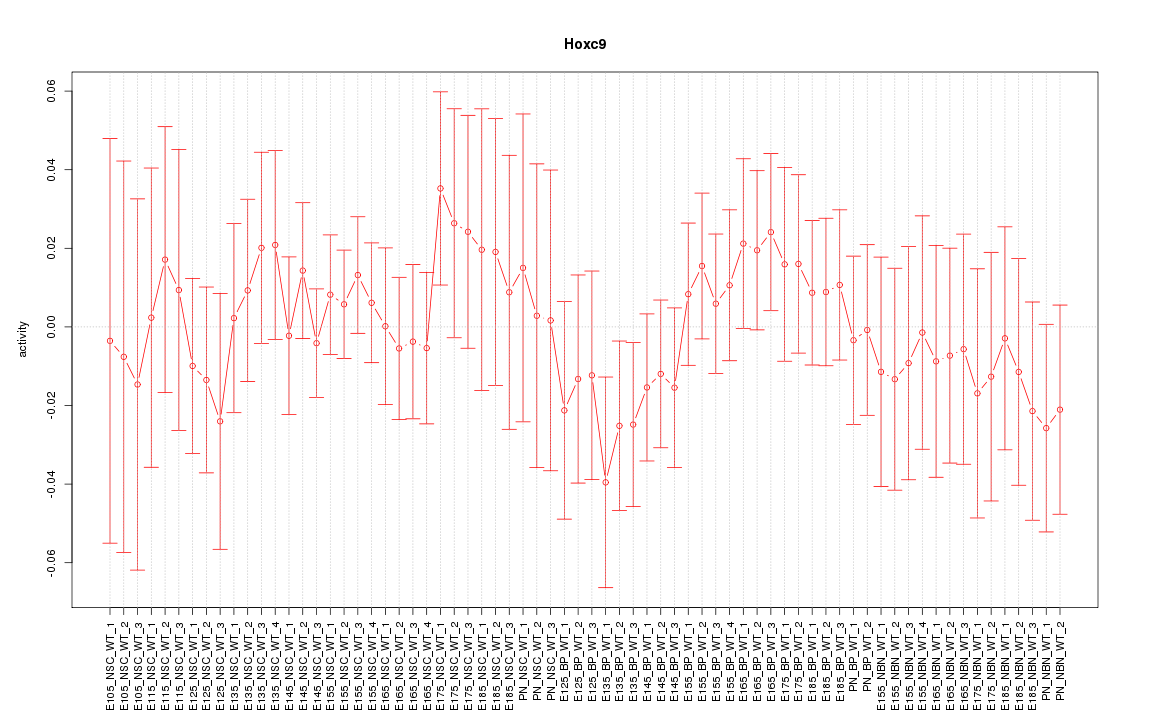
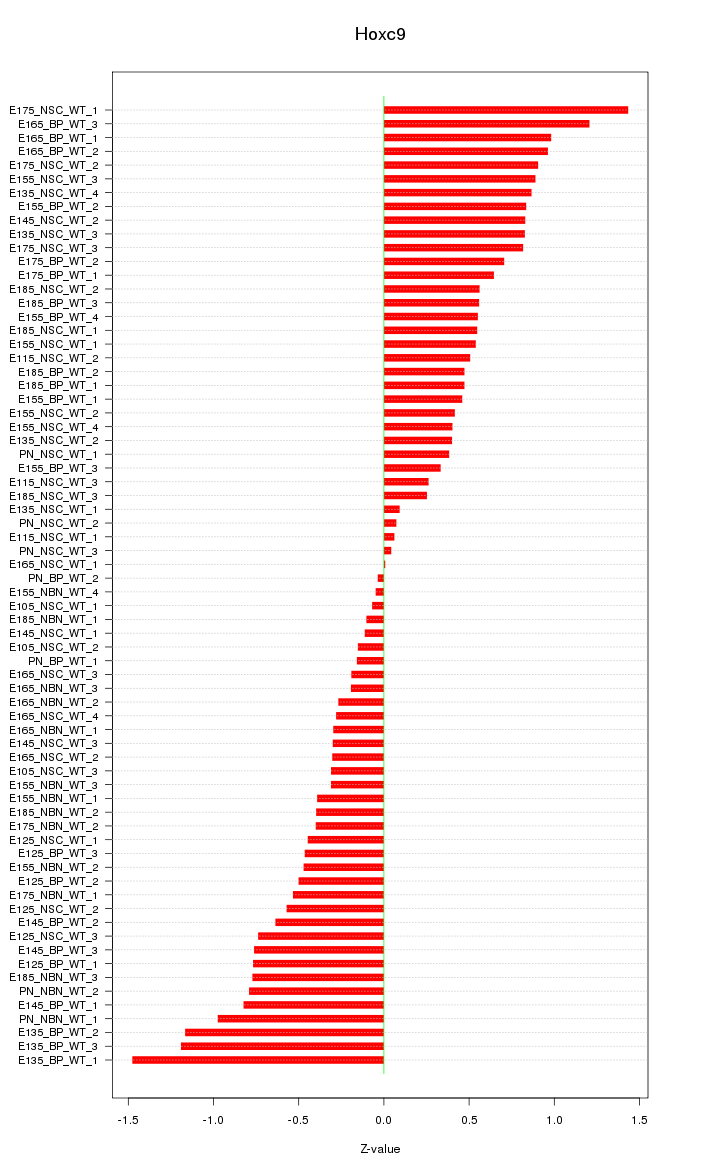
Motif ID: Hoxc9
Z-value: 0.626
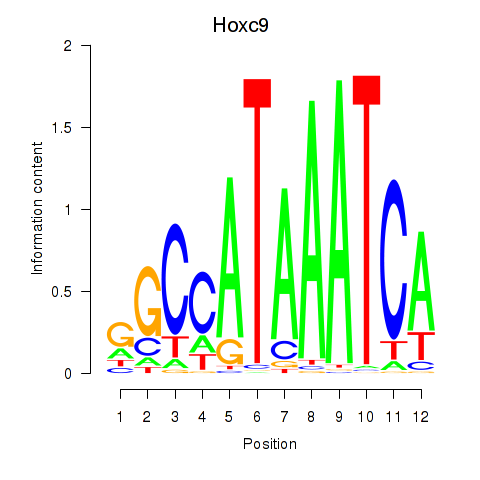
Transcription factors associated with Hoxc9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxc9 | ENSMUSG00000036139.6 | Hoxc9 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxc9 | mm10_v2_chr15_+_102977032_102977032 | 0.15 | 2.1e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0003219 | atrioventricular node development(GO:0003162) cardiac right ventricle formation(GO:0003219) |
| 1.0 | 3.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.5 | 2.1 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.5 | 2.5 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.5 | 4.0 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.5 | 2.7 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.4 | 1.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.4 | 4.6 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.4 | 1.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.2 | 2.3 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.2 | 4.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.2 | 4.8 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 | 1.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 2.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.2 | 4.2 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.2 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.4 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.1 | 0.6 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.1 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.2 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.1 | 0.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.1 | 0.2 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.1 | 2.6 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 1.7 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.1 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.9 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 1.8 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.0 | 0.9 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 1.0 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.0 | 5.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.5 | 2.4 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.4 | 1.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 2.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 2.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 2.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 4.2 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 1.7 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.2 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.7 | 2.7 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.4 | 1.6 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 1.7 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.3 | 1.3 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.3 | 2.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 4.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 5.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.2 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.6 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.4 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 4.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 3.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 3.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 2.6 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 2.5 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.0 | 0.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |