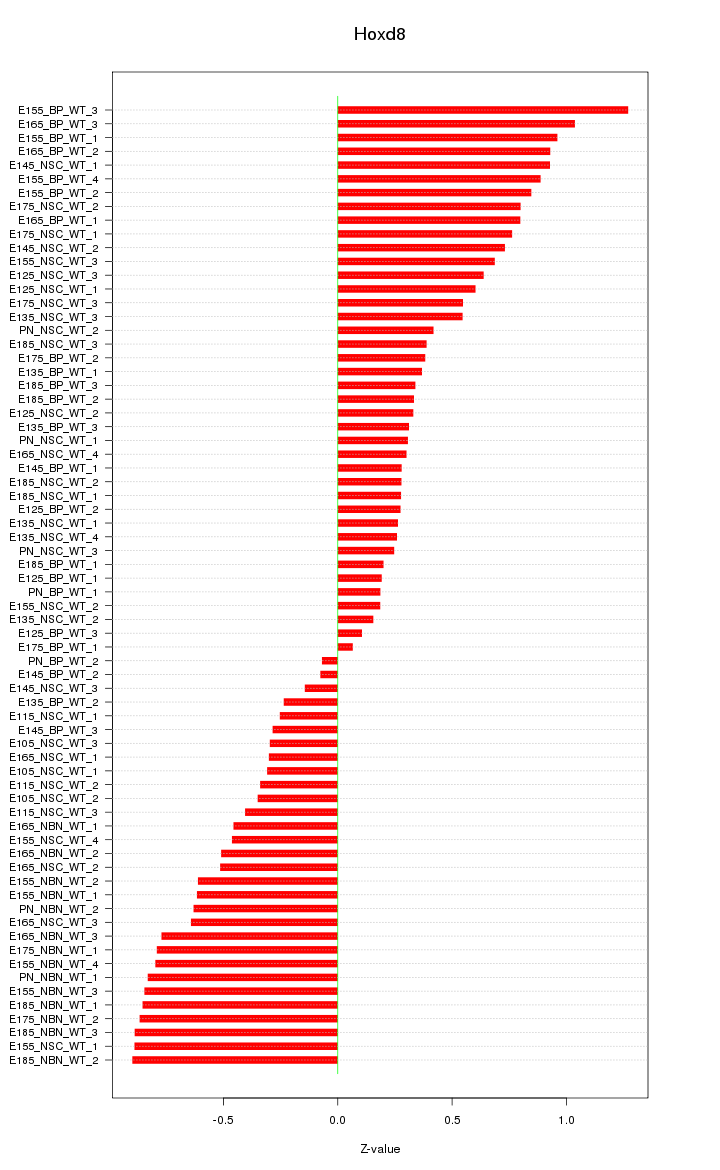
Motif ID: Hoxd8
Z-value: 0.580
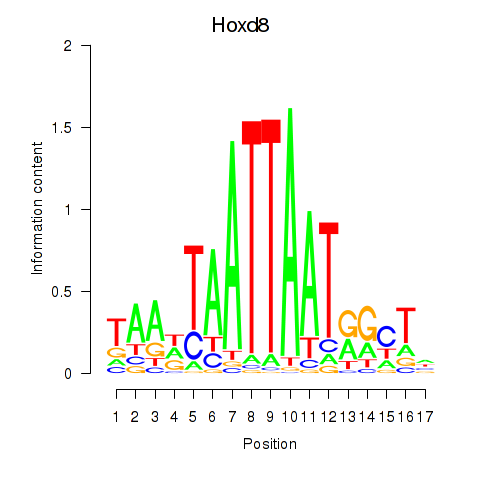
Transcription factors associated with Hoxd8:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd8 | ENSMUSG00000027102.4 | Hoxd8 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 7.6 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.7 | 9.7 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.7 | 2.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.5 | 1.6 | GO:0060489 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.5 | 2.5 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.4 | 2.5 | GO:0072539 | T-helper 17 cell differentiation(GO:0072539) |
| 0.3 | 1.3 | GO:2001045 | closure of optic fissure(GO:0061386) negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.3 | 1.6 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.3 | 0.9 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.3 | 1.3 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.3 | 1.8 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.5 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.1 | 0.5 | GO:0032911 | nerve growth factor production(GO:0032902) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.4 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:2000780 | negative regulation of double-strand break repair(GO:2000780) |
| 0.0 | 1.0 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.9 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 3.5 | GO:0021954 | central nervous system neuron development(GO:0021954) |
| 0.0 | 2.8 | GO:0006818 | hydrogen transport(GO:0006818) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.5 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 1.5 | GO:0009063 | cellular amino acid catabolic process(GO:0009063) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 2.5 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.2 | GO:0003351 | epithelial cilium movement(GO:0003351) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 4.0 | GO:0034684 | integrin alphav-beta5 complex(GO:0034684) |
| 0.4 | 1.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.4 | 9.7 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 0.9 | GO:0045251 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.0 | 2.4 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.5 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.1 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) neuregulin binding(GO:0038132) |
| 0.6 | 2.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.5 | 1.5 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.4 | 3.1 | GO:0052650 | retinal binding(GO:0016918) NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.3 | GO:0071862 | protein phosphatase type 1 activator activity(GO:0071862) |
| 0.2 | 2.5 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.0 | GO:0005402 | cation:sugar symporter activity(GO:0005402) |
| 0.1 | 10.9 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 3.5 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.5 | GO:0018741 | alkyl sulfatase activity(GO:0018741) endosulfan hemisulfate sulfatase activity(GO:0034889) endosulfan sulfate hydrolase activity(GO:0034902) |
| 0.1 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 2.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.9 | GO:0052890 | oxidoreductase activity, acting on the CH-CH group of donors, with a flavin as acceptor(GO:0052890) |
| 0.0 | 0.2 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 1.0 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.0 | 3.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.6 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 2.8 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 0.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 1.3 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 1.5 | GO:0051015 | actin filament binding(GO:0051015) |