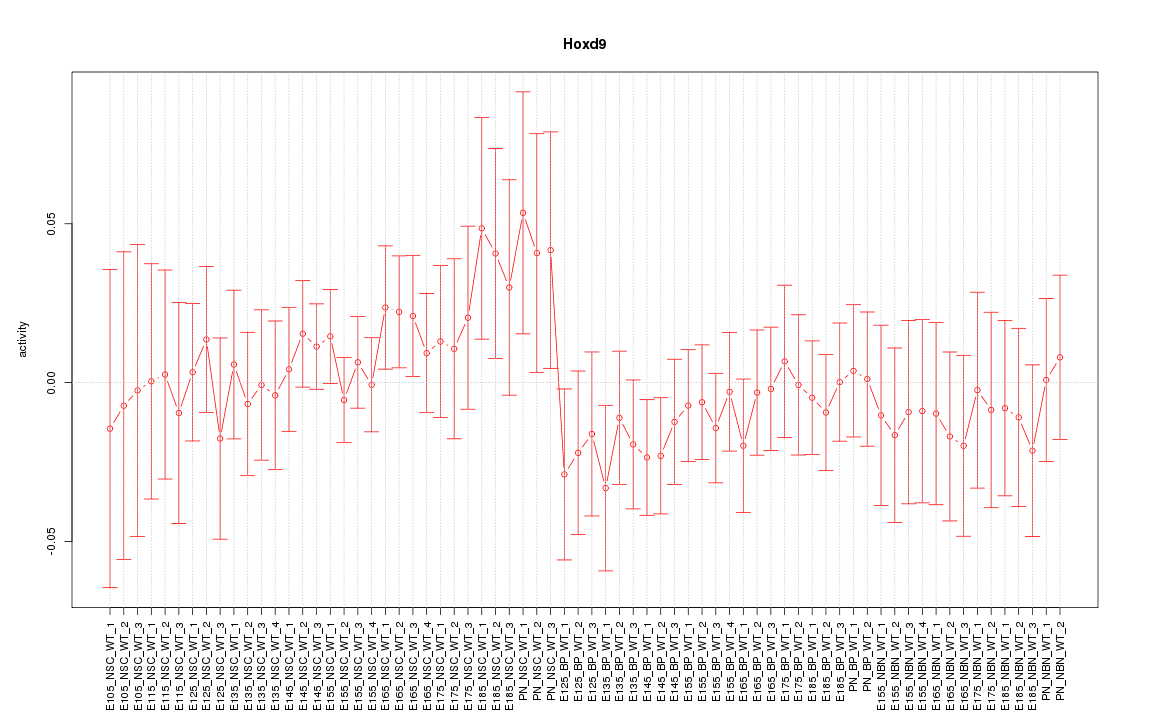
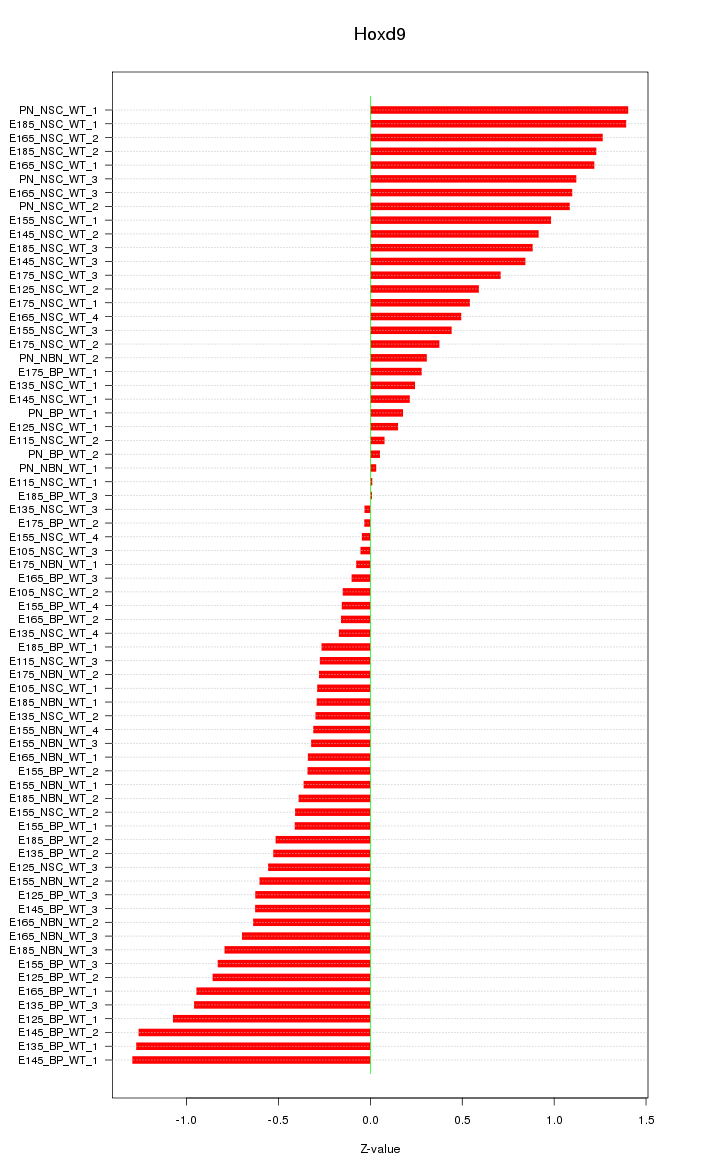
Motif ID: Hoxd9
Z-value: 0.676
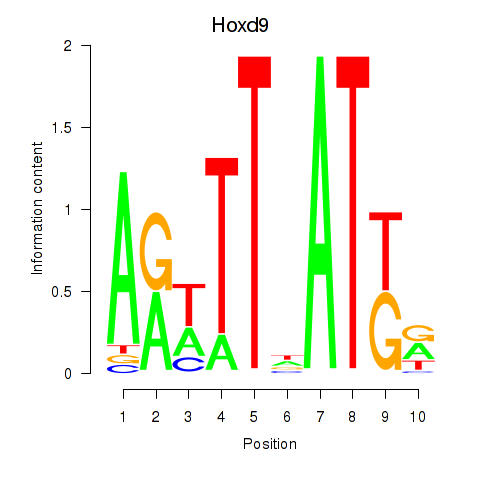
Transcription factors associated with Hoxd9:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Hoxd9 | ENSMUSG00000043342.8 | Hoxd9 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 7.9 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 1.7 | 5.0 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.1 | 8.0 | GO:0015840 | urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 1.0 | 4.0 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.9 | 10.0 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.7 | 2.2 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.7 | 2.1 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.3 | 2.4 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.3 | 9.1 | GO:0071398 | cellular response to fatty acid(GO:0071398) |
| 0.3 | 1.3 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.3 | 0.6 | GO:1904468 | negative regulation of tumor necrosis factor secretion(GO:1904468) |
| 0.3 | 3.7 | GO:0042415 | norepinephrine metabolic process(GO:0042415) |
| 0.3 | 2.3 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 1.7 | GO:0035989 | tendon development(GO:0035989) |
| 0.3 | 3.4 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.2 | 4.4 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.2 | 2.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.2 | 6.1 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 1.2 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 1.1 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 1.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.1 | 4.6 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.1 | 2.5 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 0.3 | GO:0045113 | integrin biosynthetic process(GO:0045112) regulation of integrin biosynthetic process(GO:0045113) |
| 0.1 | 1.9 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.1 | 2.1 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 4.5 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 | 0.1 | GO:2000569 | T-helper 2 cell activation(GO:0035712) positive regulation of T-helper 17 type immune response(GO:2000318) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 1.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 4.2 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 1.3 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 1.6 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.0 | 0.4 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 2.3 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.5 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.6 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.5 | 8.9 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.4 | 10.0 | GO:0002102 | podosome(GO:0002102) |
| 0.2 | 2.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.7 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.6 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.6 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.3 | GO:0071438 | NADPH oxidase complex(GO:0043020) invadopodium membrane(GO:0071438) |
| 0.1 | 5.1 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.7 | GO:0043596 | nuclear replication fork(GO:0043596) |
| 0.0 | 2.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 3.7 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.4 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 15.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 1.3 | GO:0070469 | respiratory chain(GO:0070469) |
| 0.0 | 2.3 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.0 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.7 | 5.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.3 | 8.0 | GO:0015265 | urea channel activity(GO:0015265) |
| 1.0 | 4.0 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.6 | 6.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 7.9 | GO:0017166 | dystroglycan binding(GO:0002162) vinculin binding(GO:0017166) |
| 0.4 | 2.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.3 | 2.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.3 | 4.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.3 | 2.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 1.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.2 | 1.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 9.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 1.3 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.2 | 0.5 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.2 | 2.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.2 | 3.4 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.2 | 1.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 4.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.1 | 5.5 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.3 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.1 | 3.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 3.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 1.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 1.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 1.8 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 1.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 2.0 | GO:0051015 | actin filament binding(GO:0051015) |