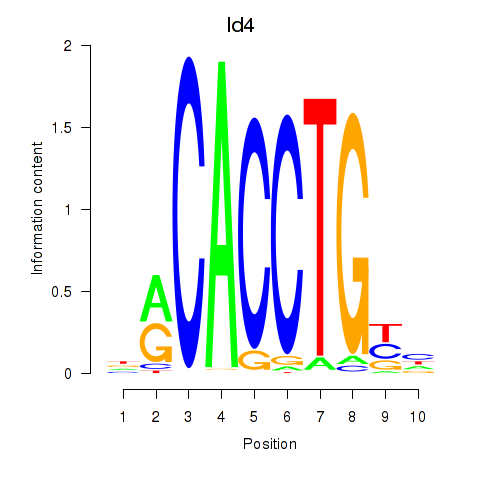
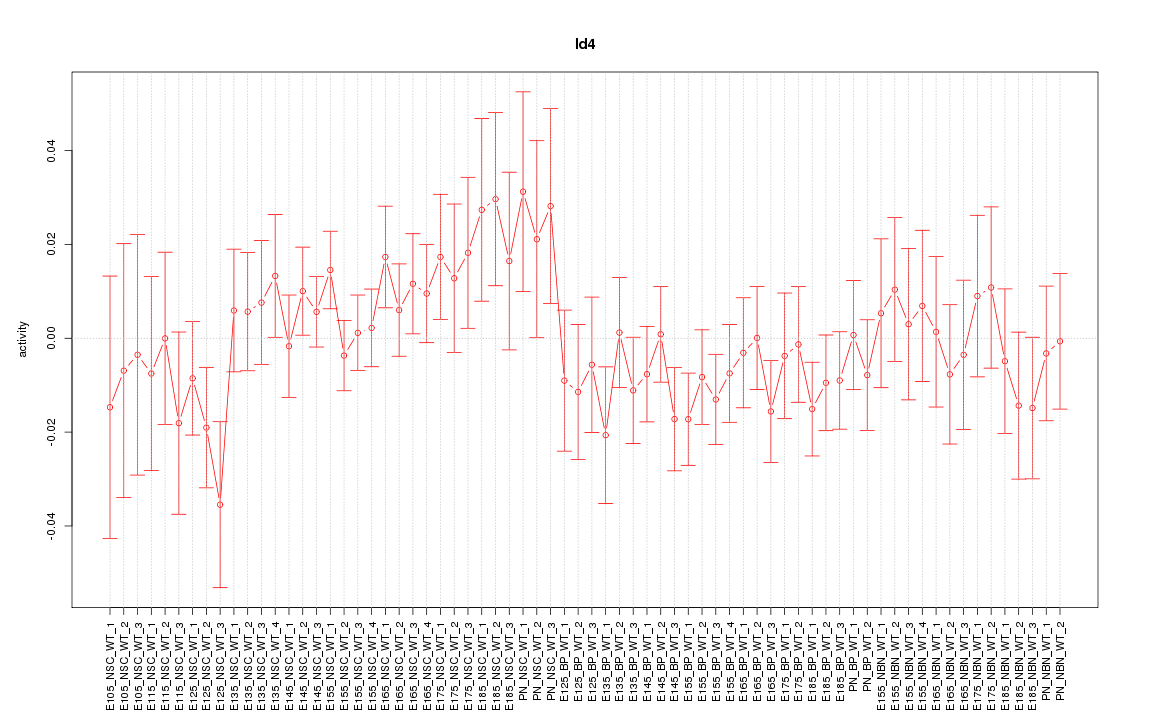
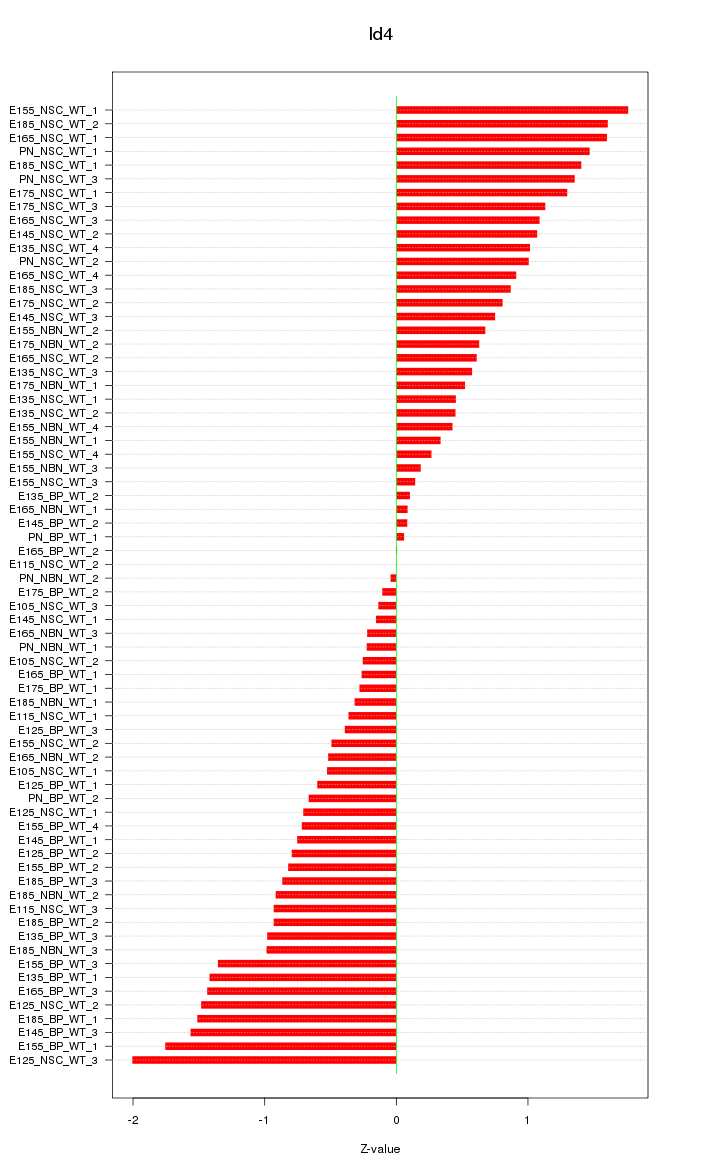
| 4.1 |
12.2 |
GO:0014826 |
vein smooth muscle contraction(GO:0014826) |
| 2.9 |
11.4 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 2.4 |
7.3 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 2.4 |
12.2 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 2.3 |
6.9 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 1.9 |
7.6 |
GO:0060032 |
notochord regression(GO:0060032) |
| 1.9 |
5.6 |
GO:0042939 |
glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.8 |
5.5 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.8 |
5.3 |
GO:0072034 |
renal vesicle induction(GO:0072034) |
| 1.7 |
5.0 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 1.7 |
1.7 |
GO:1902460 |
regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 1.6 |
6.5 |
GO:0016340 |
calcium-dependent cell-matrix adhesion(GO:0016340) |
| 1.5 |
6.0 |
GO:0019372 |
lipoxygenase pathway(GO:0019372) |
| 1.4 |
1.4 |
GO:2000823 |
regulation of androgen receptor activity(GO:2000823) |
| 1.3 |
8.8 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.2 |
7.3 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 1.2 |
5.9 |
GO:2000507 |
positive regulation of energy homeostasis(GO:2000507) |
| 1.2 |
2.3 |
GO:1903275 |
positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 1.1 |
3.3 |
GO:0010757 |
negative regulation of plasminogen activation(GO:0010757) |
| 1.1 |
3.3 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 1.0 |
3.1 |
GO:0001951 |
intestinal D-glucose absorption(GO:0001951) terminal web assembly(GO:1902896) |
| 1.0 |
4.1 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 1.0 |
5.1 |
GO:0032911 |
negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.9 |
2.8 |
GO:0072076 |
hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) negative regulation of glomerular mesangial cell proliferation(GO:0072125) negative regulation of glomerulus development(GO:0090194) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.8 |
4.1 |
GO:0075136 |
response to defenses of other organism involved in symbiotic interaction(GO:0052173) response to host defenses(GO:0052200) response to host(GO:0075136) |
| 0.8 |
3.2 |
GO:0010046 |
arginine biosynthetic process(GO:0006526) response to mycotoxin(GO:0010046) |
| 0.8 |
2.3 |
GO:1904395 |
positive regulation of receptor clustering(GO:1903911) regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.8 |
3.0 |
GO:0031581 |
hemidesmosome assembly(GO:0031581) |
| 0.7 |
2.2 |
GO:0061537 |
glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.7 |
2.1 |
GO:0036292 |
DNA rewinding(GO:0036292) |
| 0.7 |
2.1 |
GO:0071603 |
endothelial cell-cell adhesion(GO:0071603) |
| 0.7 |
2.1 |
GO:0035880 |
cell proliferation in midbrain(GO:0033278) embryonic nail plate morphogenesis(GO:0035880) |
| 0.6 |
2.5 |
GO:0046836 |
glycolipid transport(GO:0046836) |
| 0.6 |
1.8 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.6 |
4.7 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.6 |
2.9 |
GO:2000668 |
dendritic cell apoptotic process(GO:0097048) regulation of dendritic cell apoptotic process(GO:2000668) |
| 0.6 |
1.7 |
GO:2000564 |
regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) negative regulation of regulatory T cell differentiation(GO:0045590) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.6 |
5.7 |
GO:0000052 |
citrulline metabolic process(GO:0000052) |
| 0.6 |
2.3 |
GO:0070358 |
actin polymerization-dependent cell motility(GO:0070358) |
| 0.6 |
2.3 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.6 |
2.8 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.5 |
2.6 |
GO:2000189 |
positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.5 |
4.1 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.5 |
2.0 |
GO:1990379 |
lipid transport across blood brain barrier(GO:1990379) |
| 0.5 |
2.5 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.5 |
1.5 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.5 |
4.3 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.5 |
2.3 |
GO:0015871 |
choline transport(GO:0015871) |
| 0.5 |
1.4 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.5 |
0.5 |
GO:0050810 |
regulation of steroid biosynthetic process(GO:0050810) |
| 0.5 |
5.0 |
GO:0045723 |
positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.4 |
1.3 |
GO:0006682 |
galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.4 |
2.1 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 |
1.7 |
GO:2001271 |
negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.4 |
1.7 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.4 |
1.2 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.4 |
1.2 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 |
2.8 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 |
3.6 |
GO:0060662 |
tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.4 |
0.8 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.4 |
2.4 |
GO:0030913 |
paranodal junction assembly(GO:0030913) |
| 0.4 |
2.4 |
GO:0045188 |
optic nerve morphogenesis(GO:0021631) regulation of circadian sleep/wake cycle, non-REM sleep(GO:0045188) |
| 0.4 |
2.8 |
GO:0061092 |
regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.4 |
3.2 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.4 |
1.2 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.4 |
1.5 |
GO:1903181 |
regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) |
| 0.4 |
4.0 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.4 |
1.1 |
GO:0046959 |
habituation(GO:0046959) |
| 0.3 |
0.7 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.3 |
2.8 |
GO:0032261 |
purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.3 |
1.0 |
GO:0010626 |
negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.3 |
1.4 |
GO:0045054 |
constitutive secretory pathway(GO:0045054) |
| 0.3 |
1.0 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.3 |
1.3 |
GO:1903011 |
negative regulation of bone development(GO:1903011) |
| 0.3 |
0.7 |
GO:0032286 |
central nervous system myelin maintenance(GO:0032286) |
| 0.3 |
1.6 |
GO:0046601 |
positive regulation of centriole replication(GO:0046601) |
| 0.3 |
1.0 |
GO:0035106 |
operant conditioning(GO:0035106) |
| 0.3 |
3.1 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.3 |
1.6 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.3 |
3.4 |
GO:2000653 |
regulation of genetic imprinting(GO:2000653) |
| 0.3 |
0.9 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.3 |
1.5 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.3 |
7.2 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 |
0.9 |
GO:1904996 |
positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 |
4.4 |
GO:0032793 |
positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.3 |
2.0 |
GO:0045187 |
regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.3 |
1.4 |
GO:0032346 |
positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 |
1.1 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.3 |
2.5 |
GO:0032836 |
glomerular basement membrane development(GO:0032836) |
| 0.3 |
0.8 |
GO:0003162 |
atrioventricular node development(GO:0003162) |
| 0.3 |
0.8 |
GO:2001185 |
regulation of CD8-positive, alpha-beta T cell activation(GO:2001185) |
| 0.3 |
1.3 |
GO:0035590 |
purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.3 |
2.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 |
4.1 |
GO:0010640 |
regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) |
| 0.3 |
1.3 |
GO:0070779 |
D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.3 |
1.5 |
GO:0007197 |
adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 |
8.1 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.3 |
1.0 |
GO:0006432 |
phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.3 |
1.8 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.2 |
2.4 |
GO:0045741 |
positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.2 |
1.7 |
GO:0090331 |
negative regulation of platelet aggregation(GO:0090331) |
| 0.2 |
5.3 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.2 |
0.7 |
GO:0009174 |
UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.2 |
3.1 |
GO:0071257 |
cellular response to electrical stimulus(GO:0071257) |
| 0.2 |
3.3 |
GO:0042182 |
ketone catabolic process(GO:0042182) |
| 0.2 |
0.7 |
GO:0032066 |
nucleolus to nucleoplasm transport(GO:0032066) |
| 0.2 |
0.2 |
GO:0002331 |
pre-B cell allelic exclusion(GO:0002331) |
| 0.2 |
1.8 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.2 |
2.3 |
GO:0042473 |
outer ear morphogenesis(GO:0042473) |
| 0.2 |
0.9 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.2 |
0.7 |
GO:0002586 |
regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 |
2.0 |
GO:0048752 |
semicircular canal morphogenesis(GO:0048752) |
| 0.2 |
1.5 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.2 |
2.9 |
GO:0001780 |
neutrophil homeostasis(GO:0001780) negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
2.0 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.2 |
3.4 |
GO:0045672 |
positive regulation of osteoclast differentiation(GO:0045672) |
| 0.2 |
0.8 |
GO:0035166 |
post-embryonic hemopoiesis(GO:0035166) |
| 0.2 |
0.6 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.2 |
1.5 |
GO:0033563 |
dorsal/ventral axon guidance(GO:0033563) |
| 0.2 |
1.9 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.2 |
3.3 |
GO:0032060 |
bleb assembly(GO:0032060) |
| 0.2 |
1.0 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.2 |
1.8 |
GO:0014842 |
skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.2 |
0.6 |
GO:0019441 |
tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.2 |
1.0 |
GO:0006027 |
glycosaminoglycan catabolic process(GO:0006027) |
| 0.2 |
2.0 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.2 |
1.4 |
GO:1990086 |
lens fiber cell apoptotic process(GO:1990086) |
| 0.2 |
1.8 |
GO:0097070 |
ductus arteriosus closure(GO:0097070) |
| 0.2 |
2.7 |
GO:0002328 |
pro-B cell differentiation(GO:0002328) |
| 0.2 |
3.4 |
GO:0008209 |
androgen metabolic process(GO:0008209) |
| 0.2 |
0.8 |
GO:2001106 |
regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 |
0.6 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.2 |
3.7 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.2 |
0.9 |
GO:1904017 |
positive regulation of female receptivity(GO:0045925) response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.2 |
0.7 |
GO:0042219 |
cellular modified amino acid catabolic process(GO:0042219) |
| 0.2 |
0.5 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
0.7 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.2 |
0.4 |
GO:0060166 |
olfactory pit development(GO:0060166) |
| 0.2 |
1.2 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 0.2 |
1.6 |
GO:2000741 |
positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.2 |
1.6 |
GO:0051409 |
response to nitrosative stress(GO:0051409) |
| 0.2 |
0.9 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.2 |
0.7 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.2 |
1.0 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.2 |
1.0 |
GO:0030718 |
germ-line stem cell population maintenance(GO:0030718) |
| 0.2 |
1.9 |
GO:1902993 |
positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.2 |
0.2 |
GO:1902988 |
neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) |
| 0.2 |
0.5 |
GO:0051182 |
coenzyme transport(GO:0051182) |
| 0.2 |
0.9 |
GO:1903352 |
ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.2 |
0.8 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 |
0.9 |
GO:0002585 |
positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) |
| 0.2 |
0.8 |
GO:0090557 |
establishment of endothelial intestinal barrier(GO:0090557) |
| 0.2 |
0.6 |
GO:0015886 |
heme transport(GO:0015886) |
| 0.2 |
0.3 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.2 |
0.9 |
GO:0045218 |
zonula adherens maintenance(GO:0045218) |
| 0.2 |
1.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 |
1.2 |
GO:0070475 |
rRNA base methylation(GO:0070475) |
| 0.2 |
0.5 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
1.2 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.4 |
GO:0036388 |
pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 |
1.5 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
0.3 |
GO:0097051 |
establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.1 |
3.3 |
GO:0031954 |
positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 |
0.6 |
GO:0015732 |
prostaglandin transport(GO:0015732) |
| 0.1 |
4.4 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.1 |
0.5 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 |
0.4 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.1 |
0.4 |
GO:0014005 |
microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.1 |
2.6 |
GO:0095500 |
acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 |
0.4 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 |
0.4 |
GO:0034334 |
adherens junction maintenance(GO:0034334) intermediate filament bundle assembly(GO:0045110) |
| 0.1 |
0.9 |
GO:0030382 |
sperm mitochondrion organization(GO:0030382) |
| 0.1 |
1.3 |
GO:0070932 |
histone H3 deacetylation(GO:0070932) |
| 0.1 |
0.4 |
GO:2000983 |
regulation of ATP citrate synthase activity(GO:2000983) negative regulation of ATP citrate synthase activity(GO:2000984) |
| 0.1 |
2.5 |
GO:0030212 |
hyaluronan metabolic process(GO:0030212) |
| 0.1 |
1.2 |
GO:0008063 |
Toll signaling pathway(GO:0008063) |
| 0.1 |
2.0 |
GO:0016339 |
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 |
0.7 |
GO:0071285 |
cellular response to lithium ion(GO:0071285) |
| 0.1 |
0.8 |
GO:0045200 |
establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.1 |
0.7 |
GO:2000672 |
negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 |
1.7 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.1 |
1.9 |
GO:2000191 |
regulation of fatty acid transport(GO:2000191) |
| 0.1 |
0.3 |
GO:0006166 |
purine ribonucleoside salvage(GO:0006166) purine deoxyribonucleotide biosynthetic process(GO:0009153) |
| 0.1 |
0.2 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.1 |
0.4 |
GO:0045759 |
negative regulation of action potential(GO:0045759) |
| 0.1 |
0.4 |
GO:0042905 |
9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.1 |
0.2 |
GO:0009177 |
deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.1 |
0.4 |
GO:0035095 |
behavioral response to nicotine(GO:0035095) |
| 0.1 |
1.6 |
GO:0006152 |
purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 |
2.1 |
GO:0035873 |
lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 |
1.5 |
GO:0031065 |
positive regulation of histone deacetylation(GO:0031065) |
| 0.1 |
1.6 |
GO:0051044 |
positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.6 |
GO:0016255 |
attachment of GPI anchor to protein(GO:0016255) |
| 0.1 |
0.6 |
GO:0009235 |
cobalamin metabolic process(GO:0009235) |
| 0.1 |
0.2 |
GO:1901420 |
negative regulation of response to alcohol(GO:1901420) |
| 0.1 |
0.3 |
GO:0002669 |
positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) protein K29-linked ubiquitination(GO:0035519) |
| 0.1 |
7.3 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.1 |
0.8 |
GO:0014009 |
glial cell proliferation(GO:0014009) |
| 0.1 |
0.8 |
GO:0006104 |
succinyl-CoA metabolic process(GO:0006104) |
| 0.1 |
0.4 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
2.2 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.9 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 |
1.2 |
GO:0072520 |
seminiferous tubule development(GO:0072520) |
| 0.1 |
1.4 |
GO:0032968 |
positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.1 |
4.9 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 |
0.6 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.1 |
0.2 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
0.5 |
GO:0044351 |
macropinocytosis(GO:0044351) |
| 0.1 |
2.3 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.4 |
GO:0048312 |
intracellular distribution of mitochondria(GO:0048312) |
| 0.1 |
0.9 |
GO:0036037 |
CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
1.1 |
GO:0006767 |
water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 |
0.2 |
GO:1902475 |
L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 |
0.9 |
GO:0071447 |
cellular response to hydroperoxide(GO:0071447) |
| 0.1 |
0.3 |
GO:0070589 |
cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) cell wall biogenesis(GO:0042546) cell wall macromolecule metabolic process(GO:0044036) cell wall macromolecule biosynthetic process(GO:0044038) chain elongation of O-linked mannose residue(GO:0044845) cellular component macromolecule biosynthetic process(GO:0070589) cell wall organization or biogenesis(GO:0071554) |
| 0.1 |
1.3 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 |
1.7 |
GO:0033235 |
positive regulation of protein sumoylation(GO:0033235) |
| 0.1 |
1.3 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
0.6 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 |
0.2 |
GO:0048633 |
positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 |
0.9 |
GO:0006517 |
protein deglycosylation(GO:0006517) |
| 0.1 |
1.1 |
GO:0033137 |
negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.1 |
0.6 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
0.8 |
GO:0019368 |
fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 |
1.3 |
GO:0045667 |
regulation of osteoblast differentiation(GO:0045667) |
| 0.1 |
0.5 |
GO:0040016 |
embryonic cleavage(GO:0040016) |
| 0.1 |
0.7 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 |
0.8 |
GO:0010543 |
regulation of platelet activation(GO:0010543) |
| 0.1 |
0.1 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 |
1.3 |
GO:0034315 |
regulation of Arp2/3 complex-mediated actin nucleation(GO:0034315) |
| 0.1 |
0.5 |
GO:0050965 |
sensory perception of temperature stimulus(GO:0050951) detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.1 |
0.8 |
GO:0039702 |
viral budding via host ESCRT complex(GO:0039702) |
| 0.1 |
0.2 |
GO:1903251 |
multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.1 |
0.3 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.1 |
0.4 |
GO:0019532 |
oxalate transport(GO:0019532) |
| 0.1 |
1.3 |
GO:0035036 |
binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.1 |
0.6 |
GO:0030903 |
notochord development(GO:0030903) |
| 0.1 |
1.1 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
0.6 |
GO:0034773 |
histone H4-K20 trimethylation(GO:0034773) |
| 0.1 |
0.6 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
0.2 |
GO:0051660 |
cortical microtubule organization(GO:0043622) establishment of centrosome localization(GO:0051660) |
| 0.1 |
2.4 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.4 |
GO:0006560 |
proline metabolic process(GO:0006560) |
| 0.1 |
0.5 |
GO:0033169 |
histone H3-K9 demethylation(GO:0033169) regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.2 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 |
0.5 |
GO:0033690 |
positive regulation of osteoblast proliferation(GO:0033690) |
| 0.1 |
0.9 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.1 |
GO:0002924 |
negative regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002924) |
| 0.0 |
2.3 |
GO:0030835 |
negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 |
0.4 |
GO:0033005 |
positive regulation of mast cell activation(GO:0033005) positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 |
0.9 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.9 |
GO:0006182 |
cGMP biosynthetic process(GO:0006182) |
| 0.0 |
1.1 |
GO:0021522 |
spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 |
0.2 |
GO:0060770 |
negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 |
1.0 |
GO:0001649 |
osteoblast differentiation(GO:0001649) |
| 0.0 |
0.6 |
GO:0071281 |
cellular response to iron ion(GO:0071281) |
| 0.0 |
2.5 |
GO:0003073 |
regulation of systemic arterial blood pressure(GO:0003073) |
| 0.0 |
2.4 |
GO:0006661 |
phosphatidylinositol biosynthetic process(GO:0006661) |
| 0.0 |
0.7 |
GO:0042347 |
negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 |
0.8 |
GO:0045760 |
positive regulation of action potential(GO:0045760) |
| 0.0 |
0.6 |
GO:0001913 |
T cell mediated cytotoxicity(GO:0001913) |
| 0.0 |
0.6 |
GO:0007216 |
G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 |
0.3 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.0 |
0.2 |
GO:0019287 |
isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 |
0.8 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.0 |
0.6 |
GO:0014887 |
muscle hypertrophy in response to stress(GO:0003299) cardiac muscle adaptation(GO:0014887) cardiac muscle hypertrophy in response to stress(GO:0014898) |
| 0.0 |
0.4 |
GO:0071578 |
zinc II ion transmembrane import(GO:0071578) |
| 0.0 |
0.2 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 |
0.4 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.5 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.0 |
0.1 |
GO:0010571 |
positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 |
0.4 |
GO:1990440 |
positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 |
0.3 |
GO:0015074 |
DNA integration(GO:0015074) |
| 0.0 |
0.5 |
GO:0048286 |
lung alveolus development(GO:0048286) |
| 0.0 |
0.7 |
GO:2000001 |
regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 |
1.3 |
GO:0006730 |
one-carbon metabolic process(GO:0006730) |
| 0.0 |
0.1 |
GO:0071985 |
multivesicular body sorting pathway(GO:0071985) |
| 0.0 |
0.3 |
GO:2000766 |
negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 |
0.6 |
GO:0003417 |
growth plate cartilage development(GO:0003417) |
| 0.0 |
1.1 |
GO:0061001 |
regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 |
1.6 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.0 |
0.7 |
GO:0032933 |
response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 |
1.0 |
GO:0046889 |
positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 |
0.1 |
GO:0097264 |
self proteolysis(GO:0097264) |
| 0.0 |
0.3 |
GO:0016446 |
somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 |
0.8 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.0 |
0.2 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.4 |
GO:0018027 |
peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 |
0.8 |
GO:0031365 |
N-terminal protein amino acid modification(GO:0031365) |
| 0.0 |
0.2 |
GO:0019254 |
carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 |
0.3 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.3 |
GO:0031468 |
nuclear envelope reassembly(GO:0031468) |
| 0.0 |
1.1 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
2.3 |
GO:0030832 |
regulation of actin filament length(GO:0030832) |
| 0.0 |
1.5 |
GO:0045454 |
cell redox homeostasis(GO:0045454) |
| 0.0 |
0.2 |
GO:0007250 |
activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 |
1.0 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.0 |
0.5 |
GO:0046827 |
positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 |
0.3 |
GO:0048266 |
behavioral response to pain(GO:0048266) |
| 0.0 |
0.1 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.0 |
0.5 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.0 |
0.2 |
GO:0051497 |
negative regulation of actin filament bundle assembly(GO:0032232) negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 |
0.1 |
GO:0071712 |
ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 |
0.5 |
GO:0032801 |
receptor catabolic process(GO:0032801) |
| 0.0 |
0.8 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.0 |
0.3 |
GO:0060999 |
positive regulation of dendritic spine development(GO:0060999) |
| 0.0 |
0.3 |
GO:0048636 |
positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 |
0.1 |
GO:0010831 |
positive regulation of myotube differentiation(GO:0010831) |
| 0.0 |
0.2 |
GO:0010388 |
protein deneddylation(GO:0000338) cullin deneddylation(GO:0010388) |
| 0.0 |
0.1 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 |
0.2 |
GO:0010762 |
regulation of fibroblast migration(GO:0010762) |
| 0.0 |
0.0 |
GO:0001306 |
age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 |
0.3 |
GO:0006368 |
transcription elongation from RNA polymerase II promoter(GO:0006368) |
| 0.0 |
0.5 |
GO:0051893 |
regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.0 |
0.3 |
GO:0051973 |
positive regulation of telomerase activity(GO:0051973) |
| 0.0 |
0.5 |
GO:0000462 |
maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 |
0.1 |
GO:0001867 |
complement activation, lectin pathway(GO:0001867) |
| 0.0 |
0.2 |
GO:0071539 |
protein localization to centrosome(GO:0071539) |
| 0.0 |
0.6 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.0 |
0.8 |
GO:0006821 |
chloride transport(GO:0006821) |
| 0.0 |
0.1 |
GO:0061050 |
regulation of cell growth involved in cardiac muscle cell development(GO:0061050) |
| 0.0 |
0.3 |
GO:0000186 |
activation of MAPKK activity(GO:0000186) |
| 0.0 |
0.1 |
GO:0000729 |
DNA double-strand break processing(GO:0000729) |
| 0.0 |
0.2 |
GO:0001523 |
retinoid metabolic process(GO:0001523) |
| 0.0 |
0.8 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.1 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.8 |
GO:0006986 |
response to unfolded protein(GO:0006986) |
| 0.0 |
0.3 |
GO:0043113 |
receptor clustering(GO:0043113) |
| 0.0 |
0.2 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 |
0.3 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |