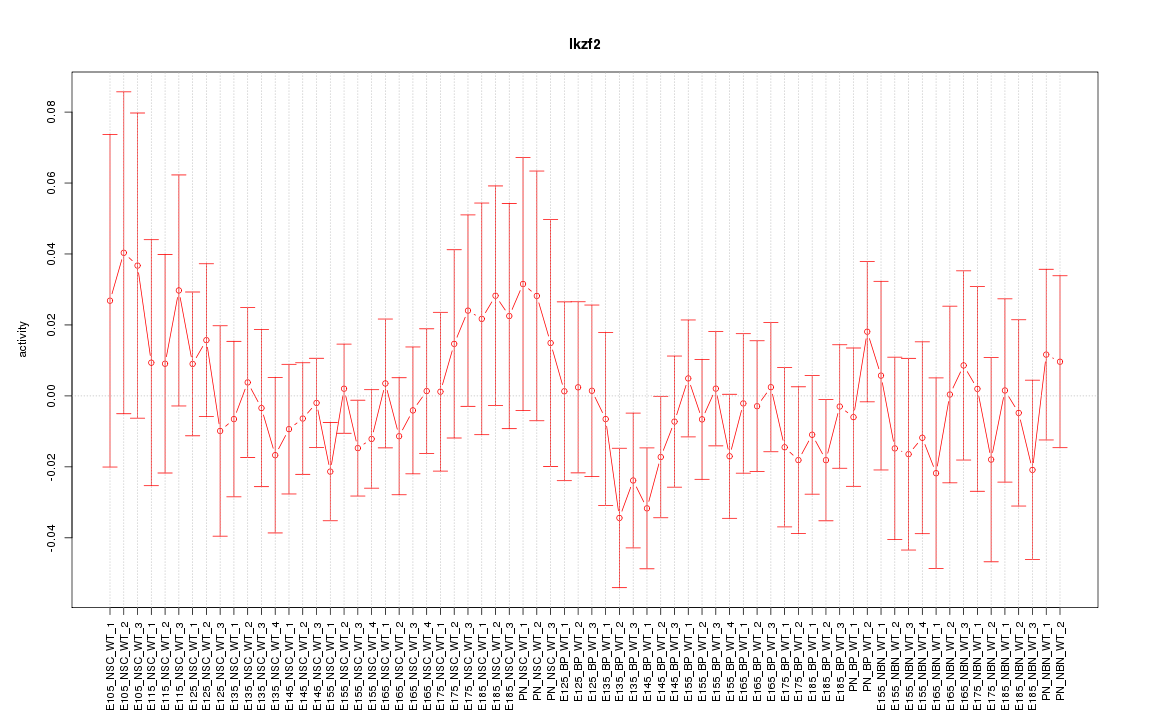
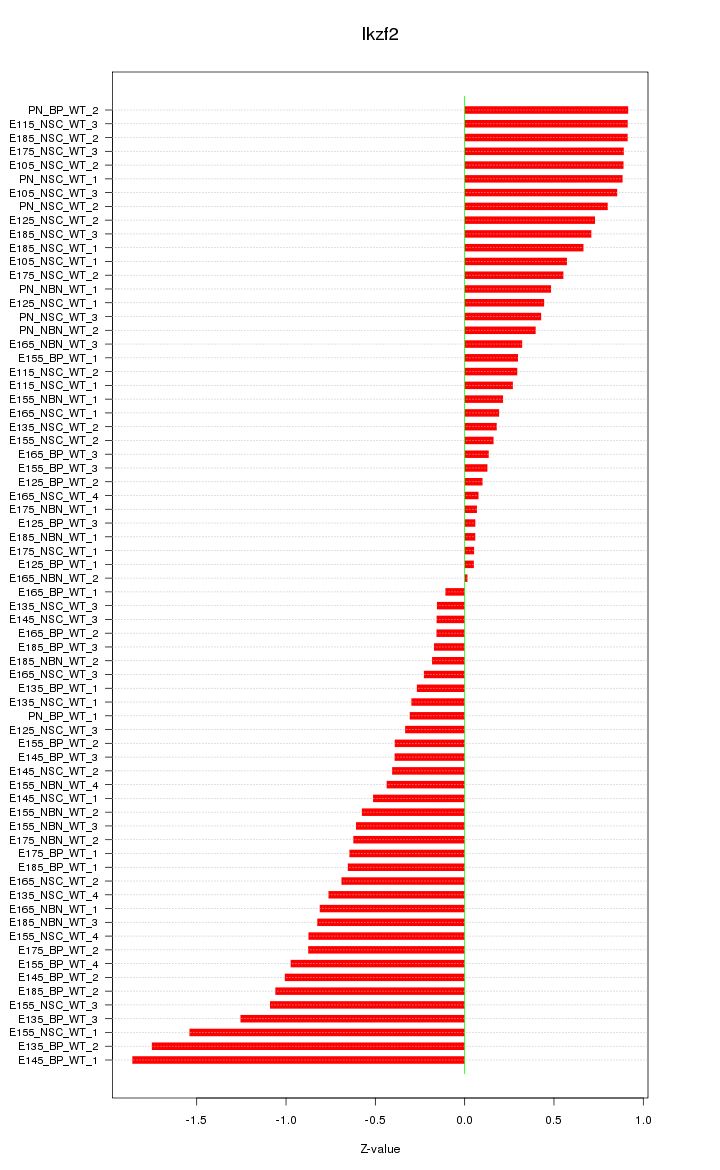
Motif ID: Ikzf2
Z-value: 0.674
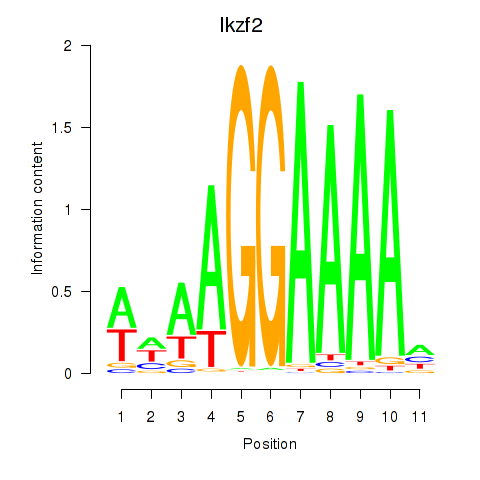
Transcription factors associated with Ikzf2:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Ikzf2 | ENSMUSG00000025997.7 | Ikzf2 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ikzf2 | mm10_v2_chr1_-_69685937_69685966 | 0.03 | 8.4e-01 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.1 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.6 | 11.2 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 1.5 | 4.5 | GO:0021557 | oculomotor nerve development(GO:0021557) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 1.4 | 8.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 1.4 | 4.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.9 | 4.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.8 | 2.5 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 2.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.7 | 7.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.7 | 2.6 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.6 | 2.4 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.6 | 3.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.6 | 4.4 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.4 | 4.3 | GO:0009404 | toxin metabolic process(GO:0009404) |
| 0.4 | 2.6 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 | 1.7 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.3 | 4.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.3 | 1.7 | GO:0019659 | lactate biosynthetic process from pyruvate(GO:0019244) lactate biosynthetic process(GO:0019249) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.3 | 0.9 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.3 | 1.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.3 | 3.6 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.3 | 0.8 | GO:0072554 | arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) endothelial cell fate specification(GO:0060847) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) positive regulation of heart induction(GO:1901321) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.3 | 1.0 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.2 | 2.3 | GO:0042447 | hormone catabolic process(GO:0042447) |
| 0.2 | 1.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.2 | 1.0 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.2 | 1.0 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 2.4 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.2 | 4.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.7 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 1.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 0.8 | GO:0090037 | positive regulation of protein kinase C signaling(GO:0090037) |
| 0.1 | 4.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.1 | 0.6 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.1 | 0.6 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.8 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.1 | 0.3 | GO:0071726 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.1 | 0.2 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 4.2 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.1 | 0.6 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 0.2 | GO:0035356 | cellular triglyceride homeostasis(GO:0035356) |
| 0.1 | 2.7 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 1.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 1.6 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 2.6 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 3.1 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 1.6 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 2.9 | GO:0098780 | mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 2.0 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 1.8 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.2 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 2.5 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 4.9 | GO:0007186 | G-protein coupled receptor signaling pathway(GO:0007186) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 1.3 | 5.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.7 | 2.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.7 | 3.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.5 | 4.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.5 | 4.5 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.4 | 2.7 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.4 | 10.0 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 0.9 | GO:0034679 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) integrin alpha9-beta1 complex(GO:0034679) |
| 0.2 | 1.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.2 | 0.8 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 1.7 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 5.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.1 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 3.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 2.1 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 0.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 4.5 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.0 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.0 | 2.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 2.9 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 4.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 2.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 2.9 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 10.0 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 2.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.9 | 4.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.7 | 11.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.5 | 2.8 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.5 | 16.7 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.4 | 4.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.4 | 1.7 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.3 | 2.3 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.3 | 1.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.3 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.9 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 2.6 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.2 | 4.4 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.2 | 4.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.7 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.8 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 3.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 5.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 1.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 6.1 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 11.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 2.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.1 | 4.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 1.1 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 1.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 5.2 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.8 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 2.6 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 2.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 1.5 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 4.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |