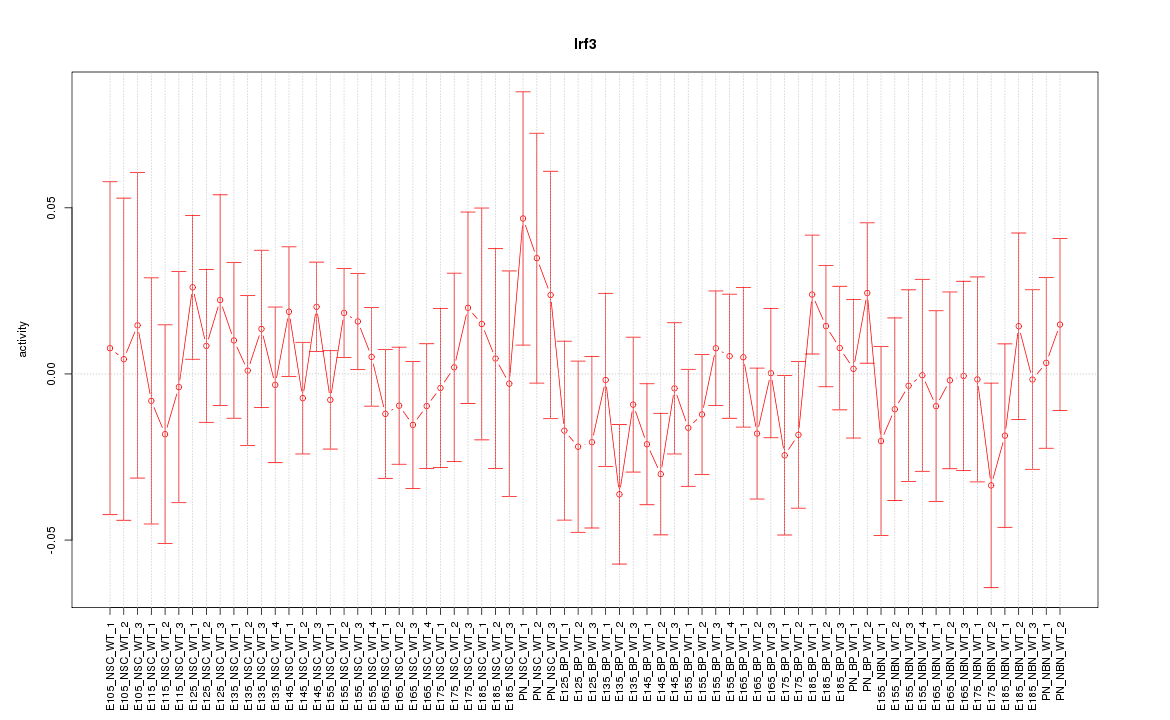
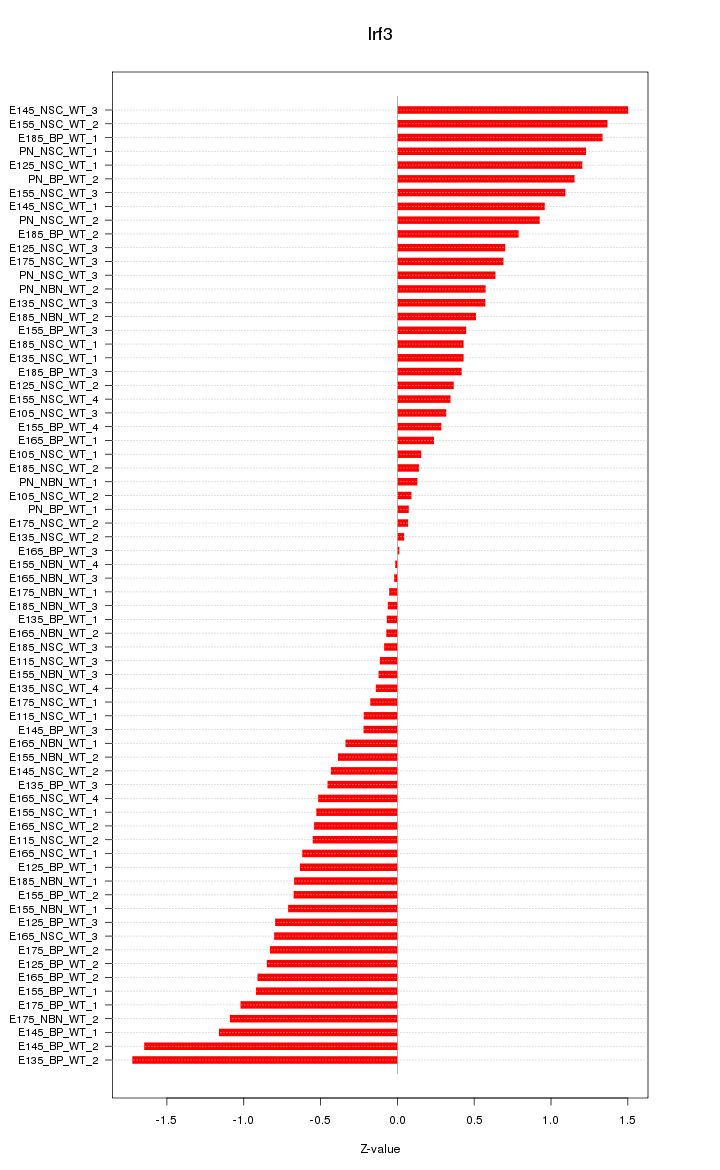
Motif ID: Irf3
Z-value: 0.710
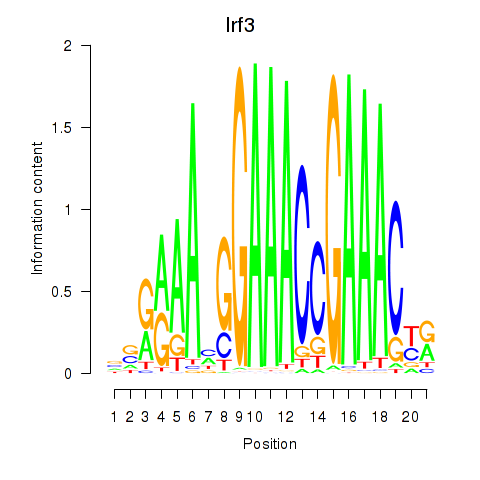
Transcription factors associated with Irf3:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Irf3 | ENSMUSG00000003184.8 | Irf3 |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf3 | mm10_v2_chr7_+_44997648_44997700 | 0.25 | 3.4e-02 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 1.0 | 4.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.9 | 6.0 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.8 | 2.4 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) musculoskeletal movement, spinal reflex action(GO:0050883) olfactory pit development(GO:0060166) |
| 0.7 | 2.6 | GO:0009597 | detection of virus(GO:0009597) |
| 0.6 | 9.3 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.4 | 2.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.4 | 1.1 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol biosynthetic process(GO:0019401) |
| 0.4 | 1.8 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 1.7 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.2 | 2.7 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.2 | 1.3 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.2 | 1.0 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.2 | 5.1 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.2 | 0.5 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) |
| 0.2 | 1.2 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.1 | 0.5 | GO:0042228 | interleukin-8 biosynthetic process(GO:0042228) |
| 0.1 | 0.8 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 0.2 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
| 0.1 | 0.6 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.1 | 0.6 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 1.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 1.2 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.7 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 3.4 | GO:0048814 | regulation of dendrite morphogenesis(GO:0048814) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.5 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 2.0 | GO:0006457 | protein folding(GO:0006457) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.8 | 9.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.6 | 0.6 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.6 | 1.7 | GO:0042025 | viral replication complex(GO:0019034) host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.5 | 4.9 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.4 | 2.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.3 | 1.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.2 | 1.0 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.3 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 4.7 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 2.9 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.2 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.3 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 2.6 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 4.2 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 2.0 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.8 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 1.0 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 2.8 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 2.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.4 | 1.7 | GO:0047844 | deoxycytidine deaminase activity(GO:0047844) |
| 0.4 | 8.1 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.4 | 2.9 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.3 | 1.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.3 | 0.8 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 2.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 2.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 0.6 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 1.0 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 4.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 2.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) E-box binding(GO:0070888) |
| 0.0 | 3.2 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 1.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 6.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.0 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 3.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.0 | 3.1 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 1.0 | GO:0051082 | unfolded protein binding(GO:0051082) |