Motif ID: Meox1
Z-value: 0.810
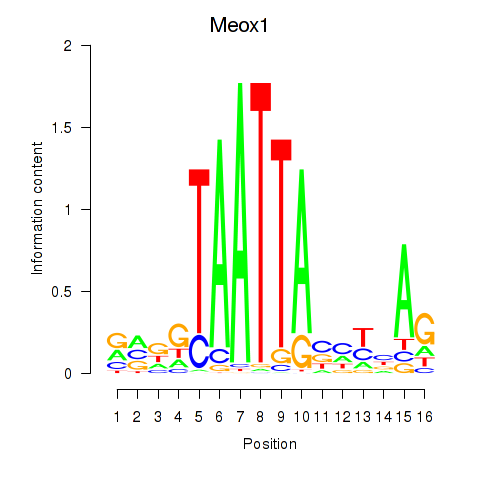
Transcription factors associated with Meox1:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Meox1 | ENSMUSG00000001493.9 | Meox1 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.9 | 27.6 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 2.7 | 8.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 2.6 | 7.9 | GO:1904395 | regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904393) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 1.7 | 11.7 | GO:0019532 | oxalate transport(GO:0019532) |
| 1.5 | 5.9 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 1.2 | 3.6 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.8 | 2.5 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.7 | 2.9 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.7 | 2.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.7 | 4.8 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.6 | 4.7 | GO:2000809 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.3 | 3.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.3 | 1.5 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.3 | 1.7 | GO:1903056 | regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.3 | 1.4 | GO:0015819 | lysine transport(GO:0015819) |
| 0.3 | 3.9 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.3 | 1.3 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.2 | 0.9 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.2 | 0.8 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 5.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.2 | 13.6 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 2.7 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 3.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.1 | 1.3 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 0.5 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) |
| 0.1 | 0.4 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.3 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 1.5 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.4 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.3 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.5 | GO:0002553 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 2.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.1 | 0.2 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.1 | 2.2 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 5.8 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 6.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 1.3 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.5 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:1904956 | response to heparin(GO:0071503) cellular response to heparin(GO:0071504) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 1.3 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.7 | 27.6 | GO:0043205 | fibril(GO:0043205) |
| 0.9 | 4.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 2.4 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.3 | 2.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.2 | 0.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.1 | 1.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 2.2 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.1 | 0.5 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 3.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 7.9 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.1 | 2.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 3.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 11.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 1.6 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 3.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 3.1 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 4.2 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 7.8 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 3.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.3 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 1.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.5 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 2.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.9 | GO:0005903 | brush border(GO:0005903) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 27.6 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.7 | 8.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 2.6 | 7.9 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 1.5 | 5.9 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 1.1 | 11.7 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.8 | 2.5 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.8 | 4.7 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.7 | 3.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.6 | 4.8 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 3.6 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled adenosine receptor activity(GO:0001609) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.4 | 1.3 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.4 | 13.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 2.9 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.3 | 3.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.3 | 13.1 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.2 | 2.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.2 | 1.4 | GO:0015189 | L-ornithine transmembrane transporter activity(GO:0000064) arginine transmembrane transporter activity(GO:0015181) L-lysine transmembrane transporter activity(GO:0015189) |
| 0.2 | 7.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.3 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.9 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 2.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.1 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 6.5 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.4 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.1 | GO:0044824 | integrase activity(GO:0008907) T/G mismatch-specific endonuclease activity(GO:0043765) retroviral integrase activity(GO:0044823) retroviral 3' processing activity(GO:0044824) |
| 0.0 | 1.3 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 1.3 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.5 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.4 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.9 | GO:0008276 | protein methyltransferase activity(GO:0008276) |
| 0.0 | 1.5 | GO:0030165 | PDZ domain binding(GO:0030165) |





