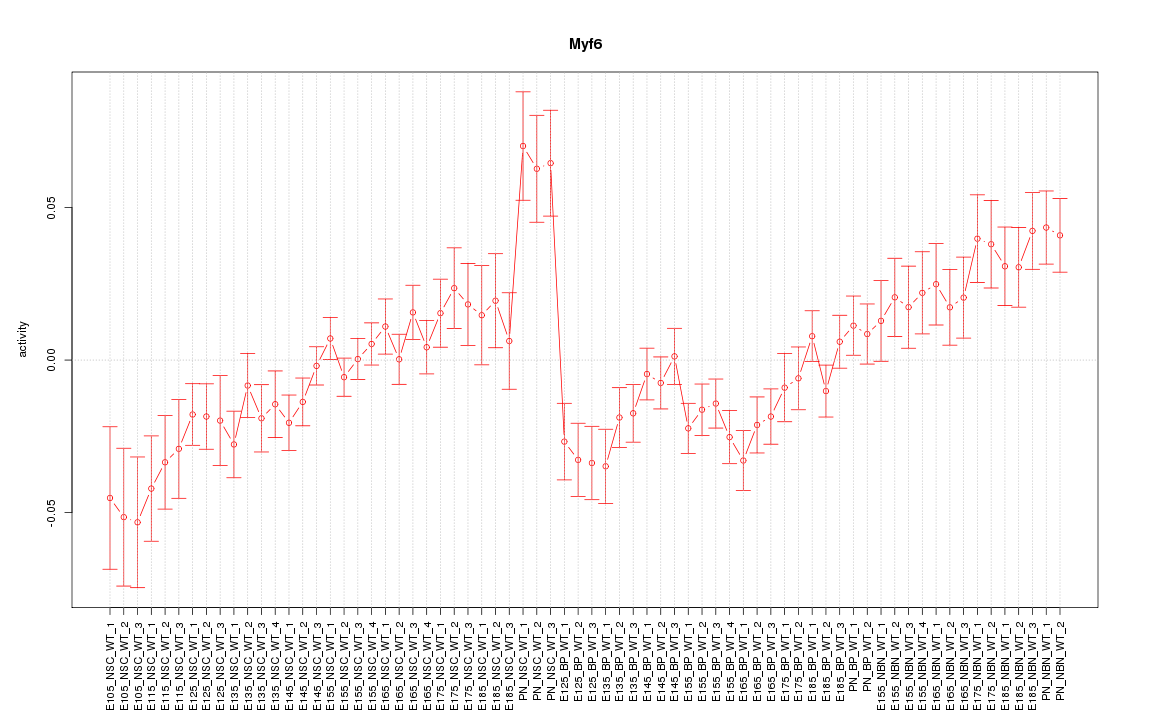
Motif ID: Myf6
Z-value: 1.997
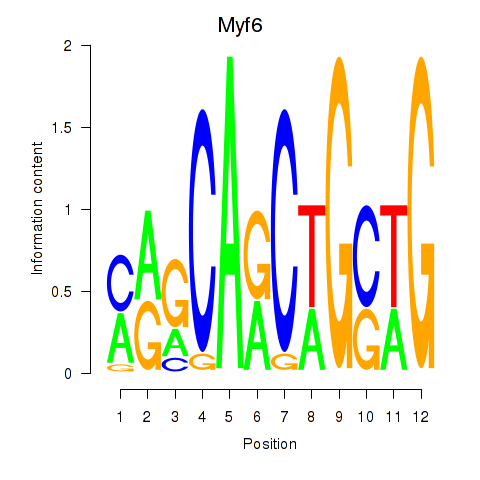
Transcription factors associated with Myf6:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Myf6 | ENSMUSG00000035923.3 | Myf6 |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.8 | 14.3 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 4.3 | 13.0 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 4.3 | 17.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 3.5 | 10.6 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 3.4 | 10.3 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 3.4 | 16.8 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 3.3 | 9.9 | GO:0046959 | habituation(GO:0046959) |
| 3.3 | 16.3 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 3.2 | 38.7 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 3.1 | 49.8 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 2.6 | 15.5 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 2.5 | 7.4 | GO:0042939 | renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 2.4 | 7.3 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.3 | 9.4 | GO:1903587 | regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 2.2 | 15.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 2.2 | 6.6 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 2.1 | 4.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 2.0 | 5.9 | GO:0097212 | lysosomal membrane organization(GO:0097212) |
| 1.9 | 5.7 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 1.8 | 7.3 | GO:0015871 | choline transport(GO:0015871) |
| 1.8 | 5.4 | GO:2000040 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 1.8 | 5.4 | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.7 | 12.2 | GO:0034047 | regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.7 | 8.4 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.7 | 5.0 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 1.6 | 4.8 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 1.6 | 6.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) lateral motor column neuron migration(GO:0097477) |
| 1.6 | 9.4 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 1.5 | 9.2 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 1.5 | 6.0 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.4 | 4.3 | GO:0003195 | tricuspid valve development(GO:0003175) tricuspid valve morphogenesis(GO:0003186) tricuspid valve formation(GO:0003195) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 1.4 | 4.3 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 1.3 | 11.8 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.3 | 6.4 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 1.3 | 5.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 1.3 | 2.5 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 1.3 | 10.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 1.2 | 1.2 | GO:0006971 | hypotonic response(GO:0006971) |
| 1.2 | 3.5 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 1.2 | 3.5 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 1.1 | 6.8 | GO:0015879 | carnitine transport(GO:0015879) |
| 1.1 | 4.5 | GO:0097393 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 1.1 | 10.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 1.1 | 5.5 | GO:2000587 | regulation of phospholipase A2 activity(GO:0032429) regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 1.1 | 10.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 1.1 | 4.3 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 1.1 | 4.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 1.0 | 10.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 1.0 | 11.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 1.0 | 3.9 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 1.0 | 26.1 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 1.0 | 3.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 1.0 | 2.9 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.9 | 10.2 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.9 | 8.3 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.9 | 4.6 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.9 | 2.7 | GO:0002578 | negative regulation of antigen processing and presentation(GO:0002578) |
| 0.9 | 3.6 | GO:0002606 | dendritic cell antigen processing and presentation(GO:0002468) regulation of dendritic cell antigen processing and presentation(GO:0002604) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.9 | 14.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.9 | 2.6 | GO:1902358 | sulfate transmembrane transport(GO:1902358) |
| 0.8 | 3.4 | GO:1904451 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.8 | 2.5 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.8 | 1.6 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.8 | 3.2 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.8 | 9.9 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.8 | 2.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.7 | 3.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.7 | 4.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.7 | 6.5 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.7 | 5.9 | GO:0071435 | potassium ion export(GO:0071435) |
| 0.7 | 13.8 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.6 | 2.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 11.9 | GO:2000060 | positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000060) |
| 0.6 | 1.9 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.6 | 0.6 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.6 | 9.2 | GO:0001964 | startle response(GO:0001964) |
| 0.6 | 4.8 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.6 | 9.6 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.6 | 9.0 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.6 | 1.8 | GO:0072070 | loop of Henle development(GO:0072070) metanephric loop of Henle development(GO:0072236) |
| 0.6 | 25.0 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.6 | 4.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.6 | 2.3 | GO:0050923 | glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) regulation of negative chemotaxis(GO:0050923) |
| 0.6 | 5.6 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.6 | 1.7 | GO:0061043 | regulation of vascular wound healing(GO:0061043) |
| 0.5 | 11.0 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.5 | 10.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.5 | 4.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.5 | 2.0 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.5 | 3.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.5 | 8.2 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.5 | 8.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.5 | 12.7 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.5 | 3.6 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.4 | 1.3 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.4 | 0.4 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.4 | 3.8 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.4 | 2.9 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.4 | 1.7 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.4 | 7.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.4 | 2.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.4 | 1.6 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.4 | 4.9 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.4 | 2.0 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.4 | 1.2 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.4 | 3.4 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.4 | 3.0 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.4 | 1.9 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 4.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.4 | 0.7 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.4 | 1.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.4 | 1.5 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.4 | 3.3 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 | 1.4 | GO:0050913 | sensory perception of bitter taste(GO:0050913) |
| 0.4 | 1.4 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.4 | 2.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.4 | 2.8 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.3 | 11.0 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.3 | 2.3 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.3 | 2.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.3 | 44.0 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.3 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.3 | 1.6 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.3 | 13.3 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.3 | 1.3 | GO:0051708 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.3 | 1.6 | GO:0032055 | negative regulation of translation in response to stress(GO:0032055) |
| 0.3 | 2.8 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.3 | 1.6 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 1.2 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.3 | 2.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.3 | 4.7 | GO:0001553 | luteinization(GO:0001553) |
| 0.3 | 0.6 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.3 | 1.9 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.3 | 3.9 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 1.5 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.3 | 1.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.2 | GO:0099622 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) cardiac muscle cell membrane repolarization(GO:0099622) regulation of cardiac muscle cell membrane repolarization(GO:0099623) ventricular cardiac muscle cell membrane repolarization(GO:0099625) |
| 0.2 | 1.0 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.2 | 1.2 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.2 | 1.5 | GO:0045187 | regulation of circadian sleep/wake cycle(GO:0042749) regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.2 | 2.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.2 | 23.7 | GO:0048709 | oligodendrocyte differentiation(GO:0048709) |
| 0.2 | 0.7 | GO:0010871 | negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.2 | 0.5 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) |
| 0.2 | 14.7 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 1.6 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.2 | 1.4 | GO:1901660 | calcium ion export(GO:1901660) |
| 0.2 | 5.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.1 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.2 | 5.9 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.2 | 5.5 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.2 | 7.4 | GO:0007200 | phospholipase C-activating G-protein coupled receptor signaling pathway(GO:0007200) |
| 0.2 | 5.5 | GO:0009268 | response to pH(GO:0009268) |
| 0.2 | 1.4 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.2 | 2.5 | GO:0046519 | sphingoid metabolic process(GO:0046519) |
| 0.2 | 0.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 14.9 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.2 | 4.3 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.2 | 1.3 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.2 | 12.5 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.2 | 3.0 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.2 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.2 | 1.5 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.2 | 0.5 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.2 | 1.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.6 | GO:2001224 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) positive regulation of neuron migration(GO:2001224) |
| 0.2 | 0.9 | GO:2000172 | endoplasmic reticulum tubular network assembly(GO:0071787) regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.2 | 0.7 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.2 | 0.5 | GO:0002586 | regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002580) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.2 | 7.1 | GO:0043407 | negative regulation of MAP kinase activity(GO:0043407) |
| 0.2 | 1.7 | GO:1901409 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.2 | 2.3 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.2 | 0.8 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.2 | 1.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.2 | 5.9 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.2 | 2.6 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.2 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 1.6 | GO:0042659 | regulation of cell fate specification(GO:0042659) |
| 0.1 | 1.3 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 2.1 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.1 | 2.9 | GO:0051926 | negative regulation of calcium ion transport(GO:0051926) |
| 0.1 | 1.2 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.1 | 5.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 2.5 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 2.5 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.1 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.7 | GO:0000022 | mitotic spindle elongation(GO:0000022) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.9 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.7 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 2.4 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.1 | 0.4 | GO:0071404 | cellular response to lipoprotein particle stimulus(GO:0071402) cellular response to low-density lipoprotein particle stimulus(GO:0071404) |
| 0.1 | 1.2 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 2.4 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.1 | 3.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.1 | 10.6 | GO:0098792 | xenophagy(GO:0098792) |
| 0.1 | 1.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 1.2 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 4.4 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.1 | 0.3 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.1 | 1.0 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 4.1 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.1 | 1.3 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 5.9 | GO:0055085 | transmembrane transport(GO:0055085) |
| 0.1 | 1.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 3.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 2.8 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.1 | 0.7 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.1 | 2.1 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.1 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.1 | 0.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.2 | GO:0039530 | MDA-5 signaling pathway(GO:0039530) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.1 | 1.9 | GO:0045806 | negative regulation of endocytosis(GO:0045806) |
| 0.1 | 0.2 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.1 | 0.4 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 2.0 | GO:0030168 | platelet activation(GO:0030168) |
| 0.0 | 0.6 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 2.4 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 1.5 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.3 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.1 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.0 | 1.4 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 2.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 3.8 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) embryonic appendage morphogenesis(GO:0035113) |
| 0.0 | 2.1 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0010955 | negative regulation of protein processing(GO:0010955) negative regulation of protein maturation(GO:1903318) |
| 0.0 | 0.3 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 2.1 | GO:0015992 | proton transport(GO:0015992) |
| 0.0 | 0.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 1.1 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.0 | 0.5 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.9 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 2.2 | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043123) |
| 0.0 | 1.7 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 0.2 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.7 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.0 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 2.9 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.2 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.6 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.6 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 46.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 3.6 | 14.3 | GO:0044307 | dendritic branch(GO:0044307) |
| 2.9 | 11.5 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 2.8 | 2.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 2.2 | 15.5 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 1.5 | 7.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 1.1 | 4.5 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 1.1 | 17.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.1 | 4.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 1.0 | 6.0 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 1.0 | 3.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 1.0 | 2.9 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 1.0 | 17.3 | GO:1902710 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 1.0 | 3.8 | GO:0005605 | basal lamina(GO:0005605) |
| 0.9 | 9.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.9 | 2.8 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.9 | 4.6 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.9 | 7.3 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.9 | 4.6 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.8 | 3.4 | GO:1990795 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.8 | 7.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.8 | 30.5 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.7 | 11.8 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.7 | 4.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.7 | 13.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 3.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.6 | 1.9 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.6 | 45.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.6 | 4.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.6 | 3.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.6 | 11.7 | GO:0030673 | axolemma(GO:0030673) |
| 0.5 | 2.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.5 | 3.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.5 | 2.6 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.5 | 2.6 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.5 | 2.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.5 | 2.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.5 | 1.4 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.5 | 5.7 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.4 | 1.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.4 | 9.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.4 | 3.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.4 | 1.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.4 | 2.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.4 | 1.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.4 | 11.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.3 | 2.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.3 | 5.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.3 | 9.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.3 | 0.8 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.3 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 4.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.3 | 9.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.0 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.2 | 7.9 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 10.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.2 | 3.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 26.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 2.3 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 0.7 | GO:0035867 | integrin alphav-beta5 complex(GO:0034684) alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.2 | 5.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 0.9 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 17.5 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 3.2 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.2 | 25.4 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.2 | 1.7 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 14.0 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.2 | 10.1 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 7.8 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.2 | 50.5 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.2 | 1.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 10.4 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 0.7 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.2 | 3.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.2 | 16.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 2.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 3.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 4.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 6.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.3 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 1.2 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.1 | 3.0 | GO:0030660 | Golgi-associated vesicle membrane(GO:0030660) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.7 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 2.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 1.1 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.1 | 1.8 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 3.3 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 4.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 0.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 1.5 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.7 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.1 | 1.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.1 | 3.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 2.0 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 0.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 14.8 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 3.0 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.4 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 2.2 | GO:0031301 | integral component of organelle membrane(GO:0031301) |
| 0.0 | 4.9 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 2.8 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 7.3 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 1.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.9 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.5 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.6 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 1.2 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 3.2 | GO:0005773 | vacuole(GO:0005773) |
| 0.0 | 0.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.5 | 16.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 3.8 | 3.8 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 3.6 | 10.8 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 3.3 | 16.6 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 3.0 | 9.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 2.9 | 17.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 2.4 | 26.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 2.3 | 6.8 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 2.1 | 8.4 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 2.0 | 46.5 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 1.9 | 9.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 1.8 | 7.3 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 1.7 | 10.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 1.7 | 11.8 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.6 | 11.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 1.3 | 14.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.2 | 10.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 1.2 | 4.8 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 1.2 | 3.6 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 1.2 | 4.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 1.0 | 28.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 1.0 | 17.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 1.0 | 3.8 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.9 | 13.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.9 | 5.6 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.9 | 7.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.9 | 5.5 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.8 | 2.5 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.8 | 2.4 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.8 | 10.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.8 | 38.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.8 | 3.0 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.7 | 2.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.7 | 4.8 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.7 | 6.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.6 | 4.5 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.6 | 7.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.6 | 4.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.6 | 2.5 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.6 | 5.3 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.6 | 3.5 | GO:0016917 | GABA receptor activity(GO:0016917) |
| 0.6 | 2.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.6 | 5.7 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.6 | 7.3 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.6 | 1.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.6 | 2.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.6 | 8.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.5 | 3.8 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.5 | 1.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.5 | 4.4 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.5 | 4.8 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.5 | 8.6 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.5 | 13.0 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 2.5 | GO:2001070 | starch binding(GO:2001070) |
| 0.5 | 9.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.5 | 2.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.5 | 4.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.5 | 1.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 1.4 | GO:0103116 | alpha-D-galactofuranose transporter activity(GO:0103116) |
| 0.5 | 10.3 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.5 | 1.4 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) |
| 0.4 | 10.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.4 | 4.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.4 | 6.4 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.4 | 2.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 4.3 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.4 | 6.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.4 | 3.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.4 | 2.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.4 | 1.6 | GO:0071074 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.4 | 1.2 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.4 | 1.6 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.4 | 1.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) |
| 0.4 | 2.3 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.4 | 5.0 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.3 | 3.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.3 | 5.5 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 1.0 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 1.6 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.3 | 13.7 | GO:0005179 | hormone activity(GO:0005179) |
| 0.3 | 1.3 | GO:0023029 | peptide antigen-transporting ATPase activity(GO:0015433) MHC class Ib protein binding(GO:0023029) tapasin binding(GO:0046980) |
| 0.3 | 7.5 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 3.9 | GO:0070636 | single-strand selective uracil DNA N-glycosylase activity(GO:0017065) nicotinamide riboside hydrolase activity(GO:0070635) nicotinic acid riboside hydrolase activity(GO:0070636) deoxyribonucleoside 5'-monophosphate N-glycosidase activity(GO:0070694) |
| 0.3 | 5.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.3 | 1.8 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.3 | 0.9 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.3 | 4.6 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.3 | 2.6 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.3 | 0.8 | GO:0008521 | acetyl-CoA transporter activity(GO:0008521) |
| 0.3 | 5.3 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.3 | 2.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.3 | 1.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.3 | 4.3 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.3 | 9.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.3 | 2.9 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.3 | 1.3 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) |
| 0.3 | 8.6 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.3 | 6.1 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 5.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 2.7 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 1.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 2.1 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.9 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 2.7 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 6.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.2 | 2.6 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.2 | 4.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 5.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.8 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.2 | 0.6 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) |
| 0.2 | 8.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.2 | 0.5 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.2 | 2.0 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.2 | 0.9 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 3.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 1.6 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.2 | 6.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 5.3 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 14.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 4.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 3.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 3.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 11.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 4.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 1.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 5.1 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 1.8 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.1 | 3.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.1 | 2.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.5 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.1 | 0.3 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) hedgehog family protein binding(GO:0097108) |
| 0.1 | 5.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 3.5 | GO:0005267 | potassium channel activity(GO:0005267) |
| 0.1 | 3.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.7 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.7 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 5.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.6 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 1.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 1.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 3.0 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.2 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 3.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.6 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 3.8 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.1 | 0.5 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 2.3 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.1 | 3.0 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.1 | 1.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 6.3 | GO:0016247 | channel regulator activity(GO:0016247) |
| 0.1 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 10.2 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 3.7 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 1.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.0 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.1 | 0.9 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 9.0 | GO:0005088 | Ras guanyl-nucleotide exchange factor activity(GO:0005088) |
| 0.1 | 0.4 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.1 | 0.9 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 1.3 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0034943 | acyl-CoA ligase activity(GO:0003996) 3-oxo-2-(2'-pentenyl)cyclopentane-1-octanoic acid CoA ligase activity(GO:0010435) 3-isopropenyl-6-oxoheptanoyl-CoA synthetase activity(GO:0018854) 2-oxo-delta3-4,5,5-trimethylcyclopentenylacetyl-CoA synthetase activity(GO:0018855) benzoyl acetate-CoA ligase activity(GO:0018856) 2,4-dichlorobenzoate-CoA ligase activity(GO:0018857) very long-chain fatty acid-CoA ligase activity(GO:0031957) pivalate-CoA ligase activity(GO:0034783) cyclopropanecarboxylate-CoA ligase activity(GO:0034793) adipate-CoA ligase activity(GO:0034796) citronellyl-CoA ligase activity(GO:0034823) mentha-1,3-dione-CoA ligase activity(GO:0034841) thiophene-2-carboxylate-CoA ligase activity(GO:0034842) 2,4,4-trimethylpentanoate-CoA ligase activity(GO:0034865) cis-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034942) trans-2-methyl-5-isopropylhexa-2,5-dienoate-CoA ligase activity(GO:0034943) branched-chain acyl-CoA synthetase (ADP-forming) activity(GO:0043759) aryl-CoA synthetase (ADP-forming) activity(GO:0043762) 3-hydroxypropionyl-CoA synthetase activity(GO:0043955) perillic acid:CoA ligase (ADP-forming) activity(GO:0052685) perillic acid:CoA ligase (AMP-forming) activity(GO:0052686) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (ADP-forming) activity(GO:0052687) (3R)-3-isopropenyl-6-oxoheptanoate:CoA ligase (AMP-forming) activity(GO:0052688) pristanate-CoA ligase activity(GO:0070251) malonyl-CoA synthetase activity(GO:0090409) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.3 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 4.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 1.2 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 10.4 | GO:0043774 | UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-diaminopimelate-D-alanyl-D-alanine ligase activity(GO:0008766) ribosomal S6-glutamic acid ligase activity(GO:0018169) coenzyme F420-0 gamma-glutamyl ligase activity(GO:0043773) coenzyme F420-2 alpha-glutamyl ligase activity(GO:0043774) protein-glycine ligase activity(GO:0070735) protein-glycine ligase activity, initiating(GO:0070736) protein-glycine ligase activity, elongating(GO:0070737) tubulin-glycine ligase activity(GO:0070738) |
| 0.0 | 8.7 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 2.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.6 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 1.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 2.0 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 3.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.0 | GO:0008514 | organic anion transmembrane transporter activity(GO:0008514) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.4 | GO:0005319 | lipid transporter activity(GO:0005319) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |