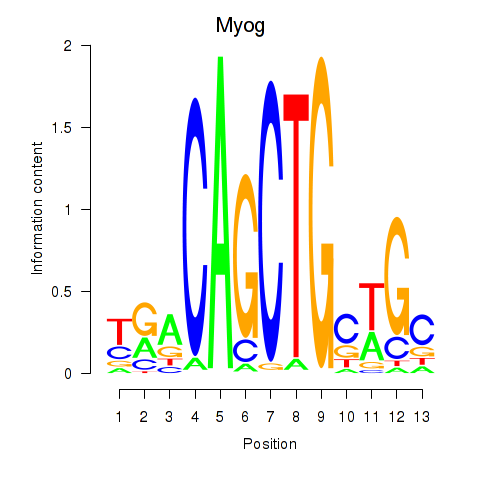
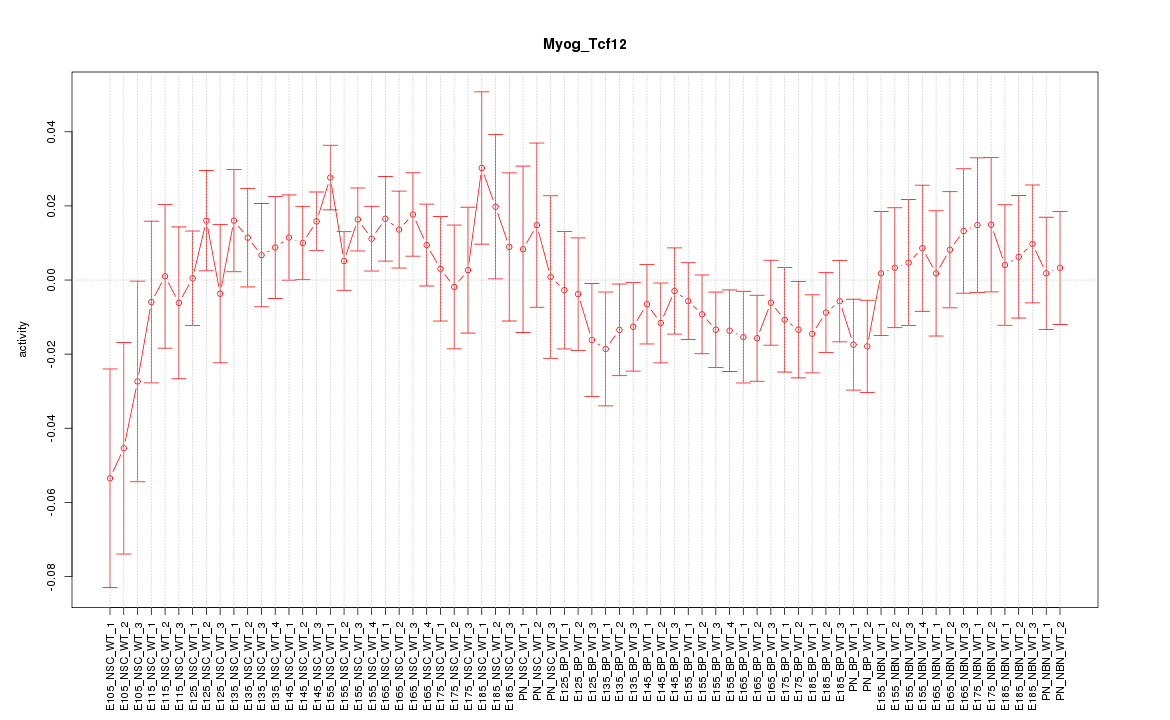
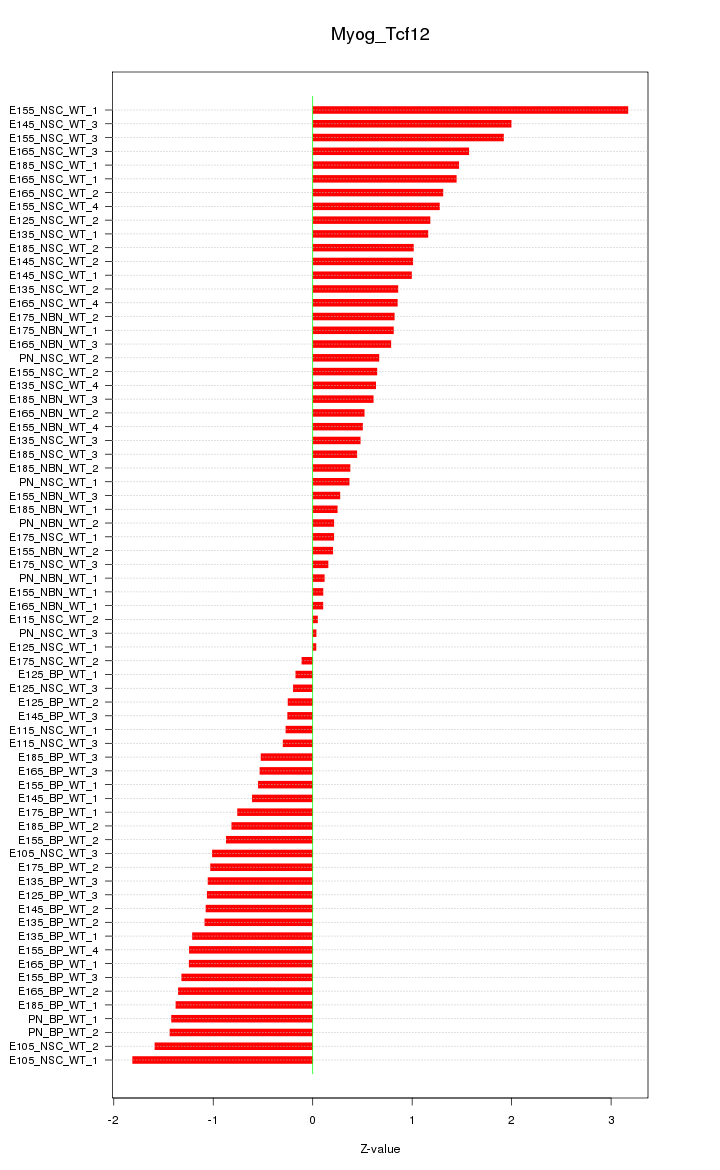
| 3.0 |
12.0 |
GO:0014012 |
peripheral nervous system axon regeneration(GO:0014012) |
| 2.6 |
7.7 |
GO:0097212 |
lysosomal membrane organization(GO:0097212) |
| 1.7 |
5.0 |
GO:0007521 |
muscle cell fate determination(GO:0007521) positive regulation of macrophage apoptotic process(GO:2000111) |
| 1.6 |
7.9 |
GO:1900085 |
negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) |
| 1.5 |
4.6 |
GO:0021893 |
cerebral cortex GABAergic interneuron fate commitment(GO:0021893) |
| 1.5 |
1.5 |
GO:0044333 |
Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 1.4 |
4.3 |
GO:0042939 |
renal sodium ion transport(GO:0003096) glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 1.4 |
9.9 |
GO:0060762 |
regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.3 |
4.0 |
GO:0050925 |
negative regulation of negative chemotaxis(GO:0050925) |
| 1.2 |
8.2 |
GO:0009449 |
gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 1.2 |
3.5 |
GO:0006601 |
creatine biosynthetic process(GO:0006601) |
| 1.1 |
7.9 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 1.1 |
3.4 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 1.1 |
4.4 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 1.0 |
9.2 |
GO:0036371 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.9 |
5.7 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.9 |
4.3 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.8 |
3.2 |
GO:0021586 |
pons maturation(GO:0021586) |
| 0.7 |
2.2 |
GO:0043465 |
regulation of fermentation(GO:0043465) negative regulation of fermentation(GO:1901003) |
| 0.7 |
4.3 |
GO:0021730 |
trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.7 |
2.1 |
GO:1904049 |
regulation of spontaneous neurotransmitter secretion(GO:1904048) negative regulation of spontaneous neurotransmitter secretion(GO:1904049) |
| 0.7 |
1.4 |
GO:0045110 |
intermediate filament bundle assembly(GO:0045110) |
| 0.7 |
4.1 |
GO:0060718 |
chorionic trophoblast cell differentiation(GO:0060718) |
| 0.7 |
2.8 |
GO:1904451 |
regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.7 |
2.7 |
GO:1903587 |
regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903587) |
| 0.7 |
2.0 |
GO:0046168 |
glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.7 |
6.6 |
GO:0051823 |
regulation of synapse structural plasticity(GO:0051823) |
| 0.6 |
5.1 |
GO:0014816 |
skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.6 |
1.9 |
GO:0035622 |
intrahepatic bile duct development(GO:0035622) |
| 0.6 |
3.1 |
GO:2000255 |
negative regulation of male germ cell proliferation(GO:2000255) |
| 0.6 |
1.9 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.6 |
1.8 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 |
2.3 |
GO:1903288 |
regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) |
| 0.6 |
4.0 |
GO:0032570 |
response to progesterone(GO:0032570) |
| 0.6 |
2.3 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.6 |
1.1 |
GO:1903275 |
positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.5 |
1.6 |
GO:0018199 |
peptidyl-glutamine modification(GO:0018199) |
| 0.5 |
1.6 |
GO:0045113 |
regulation of integrin biosynthetic process(GO:0045113) |
| 0.5 |
1.5 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.5 |
0.5 |
GO:0021966 |
corticospinal neuron axon guidance(GO:0021966) |
| 0.5 |
0.9 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.5 |
2.3 |
GO:0032423 |
regulation of mismatch repair(GO:0032423) |
| 0.5 |
1.8 |
GO:0045214 |
sarcomere organization(GO:0045214) |
| 0.4 |
1.8 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.4 |
2.2 |
GO:0036151 |
phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.4 |
0.8 |
GO:0009448 |
gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.4 |
1.6 |
GO:1902512 |
positive regulation of apoptotic DNA fragmentation(GO:1902512) positive regulation of DNA catabolic process(GO:1903626) |
| 0.4 |
1.2 |
GO:0007210 |
serotonin receptor signaling pathway(GO:0007210) |
| 0.4 |
2.0 |
GO:0014819 |
regulation of skeletal muscle contraction(GO:0014819) |
| 0.4 |
3.6 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.4 |
2.0 |
GO:0051365 |
cellular response to potassium ion starvation(GO:0051365) |
| 0.4 |
1.1 |
GO:0003349 |
epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.4 |
2.3 |
GO:0050861 |
positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.4 |
1.5 |
GO:0048739 |
cardiac muscle fiber development(GO:0048739) |
| 0.4 |
1.1 |
GO:0048686 |
regulation of sprouting of injured axon(GO:0048686) regulation of axon extension involved in regeneration(GO:0048690) |
| 0.4 |
3.9 |
GO:0002087 |
regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.4 |
2.1 |
GO:0036376 |
sodium ion export from cell(GO:0036376) |
| 0.3 |
1.4 |
GO:1900451 |
positive regulation of glutamate receptor signaling pathway(GO:1900451) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.3 |
1.4 |
GO:1900740 |
positive regulation of thymocyte apoptotic process(GO:0070245) regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.3 |
1.0 |
GO:0051542 |
elastin biosynthetic process(GO:0051542) |
| 0.3 |
3.0 |
GO:0051967 |
negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.3 |
0.9 |
GO:2000314 |
negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.3 |
0.3 |
GO:0008045 |
motor neuron axon guidance(GO:0008045) |
| 0.3 |
0.9 |
GO:0061153 |
trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.3 |
0.6 |
GO:1902174 |
positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.3 |
1.2 |
GO:0090168 |
Golgi reassembly(GO:0090168) |
| 0.3 |
12.7 |
GO:0007157 |
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.3 |
2.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.3 |
0.6 |
GO:0007158 |
neuron cell-cell adhesion(GO:0007158) |
| 0.3 |
0.8 |
GO:0046959 |
habituation(GO:0046959) |
| 0.3 |
0.3 |
GO:0060423 |
foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.3 |
2.8 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.3 |
6.1 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.3 |
0.8 |
GO:0019074 |
viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.3 |
1.1 |
GO:0051725 |
protein de-ADP-ribosylation(GO:0051725) |
| 0.3 |
1.4 |
GO:0034421 |
post-translational protein acetylation(GO:0034421) |
| 0.3 |
1.4 |
GO:0007171 |
activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.3 |
1.1 |
GO:0032227 |
negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.3 |
0.8 |
GO:0021933 |
radial glia guided migration of cerebellar granule cell(GO:0021933) |
| 0.3 |
3.9 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.3 |
0.8 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.2 |
0.7 |
GO:0010814 |
substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.2 |
1.5 |
GO:0048934 |
peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 |
1.0 |
GO:0001550 |
ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 |
0.7 |
GO:0019085 |
early viral transcription(GO:0019085) |
| 0.2 |
0.7 |
GO:1905154 |
negative regulation of membrane invagination(GO:1905154) |
| 0.2 |
1.1 |
GO:0003376 |
sphingosine-1-phosphate signaling pathway(GO:0003376) eye pigmentation(GO:0048069) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.2 |
0.4 |
GO:0060025 |
regulation of synaptic activity(GO:0060025) |
| 0.2 |
1.5 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
5.9 |
GO:0001573 |
ganglioside metabolic process(GO:0001573) |
| 0.2 |
0.2 |
GO:0015938 |
coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) |
| 0.2 |
0.4 |
GO:1902474 |
positive regulation of protein localization to synapse(GO:1902474) |
| 0.2 |
1.0 |
GO:1903347 |
negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 |
0.6 |
GO:1900060 |
negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.2 |
0.8 |
GO:0060158 |
phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.2 |
0.2 |
GO:0032696 |
negative regulation of interleukin-13 production(GO:0032696) |
| 0.2 |
1.1 |
GO:1901844 |
regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) |
| 0.2 |
8.0 |
GO:1902476 |
chloride transmembrane transport(GO:1902476) |
| 0.2 |
0.2 |
GO:0042713 |
sperm ejaculation(GO:0042713) |
| 0.2 |
0.6 |
GO:0001830 |
trophectodermal cell fate commitment(GO:0001830) |
| 0.2 |
1.4 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.2 |
1.1 |
GO:0048102 |
autophagic cell death(GO:0048102) |
| 0.2 |
1.1 |
GO:1903715 |
regulation of aerobic respiration(GO:1903715) |
| 0.2 |
0.9 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.2 |
3.4 |
GO:0008210 |
estrogen metabolic process(GO:0008210) |
| 0.2 |
1.2 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.2 |
0.8 |
GO:0051045 |
negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.2 |
0.5 |
GO:0061043 |
regulation of vascular wound healing(GO:0061043) |
| 0.2 |
0.7 |
GO:1901475 |
pyruvate transmembrane transport(GO:1901475) |
| 0.2 |
0.3 |
GO:1903998 |
regulation of eating behavior(GO:1903998) |
| 0.2 |
0.7 |
GO:0030950 |
establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.2 |
0.8 |
GO:0007021 |
tubulin complex assembly(GO:0007021) |
| 0.2 |
0.3 |
GO:0016344 |
meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.2 |
0.5 |
GO:0070634 |
transepithelial ammonium transport(GO:0070634) |
| 0.2 |
1.4 |
GO:0050966 |
detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.2 |
0.8 |
GO:2001170 |
negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 |
0.8 |
GO:0010668 |
ectodermal cell differentiation(GO:0010668) |
| 0.2 |
0.6 |
GO:0006842 |
tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.2 |
0.2 |
GO:0086042 |
cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) |
| 0.2 |
0.5 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 |
1.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.1 |
2.6 |
GO:0010763 |
positive regulation of fibroblast migration(GO:0010763) |
| 0.1 |
2.0 |
GO:0060628 |
regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 |
4.7 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
11.1 |
GO:0030032 |
lamellipodium assembly(GO:0030032) |
| 0.1 |
1.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.1 |
1.2 |
GO:0018095 |
protein polyglutamylation(GO:0018095) |
| 0.1 |
0.9 |
GO:0060923 |
cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 |
2.0 |
GO:0034501 |
protein localization to kinetochore(GO:0034501) |
| 0.1 |
1.6 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
1.2 |
GO:1902231 |
positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.1 |
0.6 |
GO:0032485 |
regulation of Ral protein signal transduction(GO:0032485) |
| 0.1 |
0.4 |
GO:0097402 |
neuroblast migration(GO:0097402) |
| 0.1 |
0.4 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.1 |
1.8 |
GO:0097120 |
receptor localization to synapse(GO:0097120) |
| 0.1 |
0.5 |
GO:0060741 |
prostate gland stromal morphogenesis(GO:0060741) |
| 0.1 |
0.4 |
GO:0018103 |
protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 |
2.7 |
GO:0045475 |
locomotor rhythm(GO:0045475) |
| 0.1 |
4.9 |
GO:0007405 |
neuroblast proliferation(GO:0007405) |
| 0.1 |
0.4 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 |
0.5 |
GO:1902237 |
positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.1 |
0.8 |
GO:1903423 |
positive regulation of synaptic vesicle recycling(GO:1903423) |
| 0.1 |
1.8 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.1 |
0.7 |
GO:0048703 |
embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.1 |
0.6 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 |
0.6 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.5 |
GO:0035507 |
regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.1 |
0.5 |
GO:0033580 |
protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.1 |
2.2 |
GO:0001964 |
startle response(GO:0001964) |
| 0.1 |
0.6 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
1.0 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 |
0.3 |
GO:1900736 |
regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 |
0.6 |
GO:1901678 |
iron coordination entity transport(GO:1901678) |
| 0.1 |
0.3 |
GO:0060912 |
cardiac cell fate specification(GO:0060912) |
| 0.1 |
1.0 |
GO:0042095 |
interferon-gamma biosynthetic process(GO:0042095) |
| 0.1 |
0.4 |
GO:0021891 |
olfactory bulb interneuron development(GO:0021891) |
| 0.1 |
0.5 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.1 |
0.6 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 |
1.6 |
GO:0070296 |
sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.1 |
0.5 |
GO:0016080 |
synaptic vesicle targeting(GO:0016080) |
| 0.1 |
1.8 |
GO:2000310 |
regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.1 |
0.4 |
GO:0014889 |
muscle atrophy(GO:0014889) |
| 0.1 |
1.0 |
GO:1900112 |
regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.1 |
0.1 |
GO:0098528 |
skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 |
1.8 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.1 |
0.9 |
GO:0071435 |
potassium ion export(GO:0071435) |
| 0.1 |
0.3 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.1 |
0.7 |
GO:0010820 |
positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 |
1.8 |
GO:0035914 |
skeletal muscle cell differentiation(GO:0035914) |
| 0.1 |
0.9 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 |
1.0 |
GO:0051103 |
DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 |
0.6 |
GO:1903025 |
regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 |
0.1 |
GO:0014016 |
neuroblast differentiation(GO:0014016) |
| 0.1 |
0.7 |
GO:0051014 |
actin filament severing(GO:0051014) |
| 0.1 |
0.3 |
GO:0009414 |
response to water deprivation(GO:0009414) |
| 0.1 |
1.1 |
GO:1900454 |
positive regulation of long term synaptic depression(GO:1900454) |
| 0.1 |
0.8 |
GO:0016925 |
protein sumoylation(GO:0016925) |
| 0.1 |
1.0 |
GO:0045687 |
positive regulation of glial cell differentiation(GO:0045687) |
| 0.1 |
1.0 |
GO:0009109 |
coenzyme catabolic process(GO:0009109) |
| 0.1 |
0.2 |
GO:0034241 |
positive regulation of macrophage fusion(GO:0034241) regulation of osteoclast proliferation(GO:0090289) |
| 0.1 |
0.3 |
GO:0050913 |
sensory perception of bitter taste(GO:0050913) |
| 0.1 |
1.6 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.1 |
0.5 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 |
0.4 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.1 |
3.9 |
GO:0000381 |
regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 |
1.1 |
GO:0050728 |
negative regulation of inflammatory response(GO:0050728) |
| 0.1 |
1.9 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.1 |
0.2 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.1 |
2.1 |
GO:0007616 |
long-term memory(GO:0007616) |
| 0.1 |
0.2 |
GO:1990481 |
mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 |
0.1 |
GO:1901642 |
purine nucleoside transmembrane transport(GO:0015860) purine-containing compound transmembrane transport(GO:0072530) nucleoside transmembrane transport(GO:1901642) |
| 0.1 |
0.3 |
GO:0072365 |
regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 |
0.1 |
GO:0021764 |
amygdala development(GO:0021764) |
| 0.1 |
1.9 |
GO:0060074 |
synapse maturation(GO:0060074) |
| 0.1 |
0.5 |
GO:0015791 |
polyol transport(GO:0015791) |
| 0.1 |
1.9 |
GO:0010842 |
retina layer formation(GO:0010842) |
| 0.1 |
0.4 |
GO:0021984 |
adenohypophysis development(GO:0021984) |
| 0.1 |
0.3 |
GO:0010499 |
proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 |
0.8 |
GO:0010866 |
regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.1 |
3.3 |
GO:0048384 |
retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 |
0.3 |
GO:0090370 |
negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 |
0.5 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.9 |
GO:1902260 |
negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 |
0.5 |
GO:0031268 |
pseudopodium organization(GO:0031268) |
| 0.1 |
5.2 |
GO:0048709 |
oligodendrocyte differentiation(GO:0048709) |
| 0.1 |
1.6 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.1 |
0.4 |
GO:0060272 |
embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.1 |
1.6 |
GO:0045806 |
negative regulation of endocytosis(GO:0045806) |
| 0.1 |
0.4 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
3.5 |
GO:0021766 |
hippocampus development(GO:0021766) |
| 0.1 |
1.1 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.1 |
0.1 |
GO:0010936 |
negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 |
0.3 |
GO:0009052 |
pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.1 |
0.2 |
GO:0045059 |
positive thymic T cell selection(GO:0045059) |
| 0.1 |
0.4 |
GO:0006620 |
posttranslational protein targeting to membrane(GO:0006620) |
| 0.1 |
0.3 |
GO:0071476 |
hypotonic response(GO:0006971) cellular hypotonic response(GO:0071476) |
| 0.0 |
0.1 |
GO:0030202 |
heparin metabolic process(GO:0030202) |
| 0.0 |
1.8 |
GO:0006890 |
retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 |
1.2 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 |
0.6 |
GO:0019262 |
N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 |
0.6 |
GO:0030574 |
collagen catabolic process(GO:0030574) |
| 0.0 |
0.8 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 |
0.1 |
GO:0036006 |
response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 |
0.4 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.5 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 |
0.2 |
GO:0021631 |
optic nerve morphogenesis(GO:0021631) |
| 0.0 |
0.8 |
GO:0031116 |
positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 |
1.6 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
0.4 |
GO:0015868 |
purine ribonucleotide transport(GO:0015868) |
| 0.0 |
0.2 |
GO:0032483 |
regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 |
1.5 |
GO:0003333 |
amino acid transmembrane transport(GO:0003333) |
| 0.0 |
0.5 |
GO:0051127 |
positive regulation of actin nucleation(GO:0051127) |
| 0.0 |
0.5 |
GO:0045217 |
cell-cell junction maintenance(GO:0045217) |
| 0.0 |
0.2 |
GO:0098903 |
regulation of membrane repolarization during action potential(GO:0098903) |
| 0.0 |
0.0 |
GO:0006701 |
progesterone biosynthetic process(GO:0006701) |
| 0.0 |
0.1 |
GO:2000620 |
regulation of histone H4-K16 acetylation(GO:2000618) positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.0 |
0.1 |
GO:0050909 |
sensory perception of taste(GO:0050909) |
| 0.0 |
0.7 |
GO:0045662 |
negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 |
0.2 |
GO:0002036 |
regulation of L-glutamate transport(GO:0002036) negative regulation of mitochondrial membrane potential(GO:0010917) |
| 0.0 |
0.2 |
GO:0034587 |
piRNA metabolic process(GO:0034587) |
| 0.0 |
0.7 |
GO:0046716 |
muscle cell cellular homeostasis(GO:0046716) |
| 0.0 |
0.3 |
GO:0014807 |
regulation of somitogenesis(GO:0014807) |
| 0.0 |
0.5 |
GO:0048488 |
synaptic vesicle endocytosis(GO:0048488) |
| 0.0 |
0.1 |
GO:0046340 |
diacylglycerol catabolic process(GO:0046340) |
| 0.0 |
0.2 |
GO:1902047 |
polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.0 |
1.5 |
GO:0007032 |
endosome organization(GO:0007032) |
| 0.0 |
0.1 |
GO:0032897 |
negative regulation of viral transcription(GO:0032897) |
| 0.0 |
0.2 |
GO:0031103 |
axon regeneration(GO:0031103) |
| 0.0 |
0.6 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.0 |
0.3 |
GO:0043045 |
DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 |
0.3 |
GO:0051574 |
positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 |
0.3 |
GO:0048199 |
vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.0 |
0.2 |
GO:0007221 |
positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 |
0.0 |
GO:0044861 |
protein transport into plasma membrane raft(GO:0044861) |
| 0.0 |
0.3 |
GO:0036158 |
outer dynein arm assembly(GO:0036158) |
| 0.0 |
1.1 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 |
0.2 |
GO:0035635 |
entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.0 |
0.4 |
GO:2000465 |
regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 |
0.4 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.3 |
GO:0007413 |
axonal fasciculation(GO:0007413) |
| 0.0 |
0.3 |
GO:1904262 |
negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 |
0.4 |
GO:0034142 |
toll-like receptor 4 signaling pathway(GO:0034142) |
| 0.0 |
0.6 |
GO:0010830 |
regulation of myotube differentiation(GO:0010830) |
| 0.0 |
1.7 |
GO:0043407 |
negative regulation of MAP kinase activity(GO:0043407) |
| 0.0 |
0.1 |
GO:0061158 |
3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 |
0.3 |
GO:0051092 |
positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 |
0.4 |
GO:1903077 |
negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 |
0.1 |
GO:0061002 |
negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 |
1.2 |
GO:0014013 |
regulation of gliogenesis(GO:0014013) |
| 0.0 |
1.6 |
GO:0098656 |
anion transmembrane transport(GO:0098656) |
| 0.0 |
1.2 |
GO:0050885 |
neuromuscular process controlling balance(GO:0050885) |
| 0.0 |
0.5 |
GO:0006970 |
response to osmotic stress(GO:0006970) |
| 0.0 |
0.1 |
GO:0010726 |
positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.0 |
0.3 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 |
0.1 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.0 |
0.2 |
GO:0014842 |
regulation of skeletal muscle satellite cell proliferation(GO:0014842) regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.0 |
0.3 |
GO:0043649 |
dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 |
0.3 |
GO:0051481 |
negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 |
0.3 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 |
0.2 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 |
0.2 |
GO:0006621 |
protein retention in ER lumen(GO:0006621) |
| 0.0 |
1.4 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 |
0.2 |
GO:0031848 |
protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 |
1.9 |
GO:0006892 |
post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.0 |
2.5 |
GO:0006342 |
chromatin silencing(GO:0006342) |
| 0.0 |
0.2 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.0 |
0.2 |
GO:0038166 |
angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 |
0.8 |
GO:0046889 |
positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 |
0.3 |
GO:0051988 |
regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 |
0.5 |
GO:0032981 |
NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 |
1.2 |
GO:0008306 |
associative learning(GO:0008306) |
| 0.0 |
0.5 |
GO:0007340 |
acrosome reaction(GO:0007340) |
| 0.0 |
0.2 |
GO:0006646 |
phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 |
0.2 |
GO:0006102 |
isocitrate metabolic process(GO:0006102) |
| 0.0 |
0.6 |
GO:0030514 |
negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 |
0.5 |
GO:0000731 |
DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 |
0.9 |
GO:0033014 |
porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.0 |
0.1 |
GO:0070166 |
enamel mineralization(GO:0070166) |
| 0.0 |
0.1 |
GO:2000643 |
positive regulation of vacuolar transport(GO:1903337) positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 |
0.3 |
GO:0090231 |
regulation of spindle checkpoint(GO:0090231) |
| 0.0 |
0.2 |
GO:0034551 |
respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 |
0.0 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 |
0.2 |
GO:0010457 |
centriole-centriole cohesion(GO:0010457) |
| 0.0 |
0.3 |
GO:0018149 |
peptide cross-linking(GO:0018149) |
| 0.0 |
0.4 |
GO:0051491 |
positive regulation of filopodium assembly(GO:0051491) |
| 0.0 |
0.3 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 |
0.6 |
GO:0034605 |
cellular response to heat(GO:0034605) |
| 0.0 |
0.9 |
GO:0019933 |
cAMP-mediated signaling(GO:0019933) |
| 0.0 |
0.1 |
GO:0045056 |
transcytosis(GO:0045056) |
| 0.0 |
0.1 |
GO:0003413 |
chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) |
| 0.0 |
0.0 |
GO:0010847 |
regulation of chromatin assembly(GO:0010847) |
| 0.0 |
0.5 |
GO:0007257 |
activation of JUN kinase activity(GO:0007257) |
| 0.0 |
0.1 |
GO:0046036 |
GTP biosynthetic process(GO:0006183) CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 |
0.2 |
GO:2001237 |
negative regulation of extrinsic apoptotic signaling pathway(GO:2001237) |
| 0.0 |
0.2 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.0 |
0.2 |
GO:0002063 |
chondrocyte development(GO:0002063) |
| 0.0 |
0.1 |
GO:0031998 |
regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 |
0.1 |
GO:0016242 |
negative regulation of macroautophagy(GO:0016242) |
| 0.0 |
1.8 |
GO:0045930 |
negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 |
0.5 |
GO:0080171 |
lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.0 |
0.2 |
GO:2000649 |
regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 |
0.1 |
GO:0051570 |
regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 |
0.5 |
GO:0035023 |
regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 |
0.3 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.1 |
GO:0050852 |
T cell receptor signaling pathway(GO:0050852) |
| 0.0 |
0.0 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.0 |
0.3 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 |
0.3 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 |
0.1 |
GO:0034244 |
negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 |
0.1 |
GO:0010606 |
positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 |
0.1 |
GO:0006614 |
SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 |
0.2 |
GO:0007064 |
mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 |
0.1 |
GO:0006152 |
purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 |
0.2 |
GO:2000398 |
regulation of T cell differentiation in thymus(GO:0033081) regulation of thymocyte aggregation(GO:2000398) |