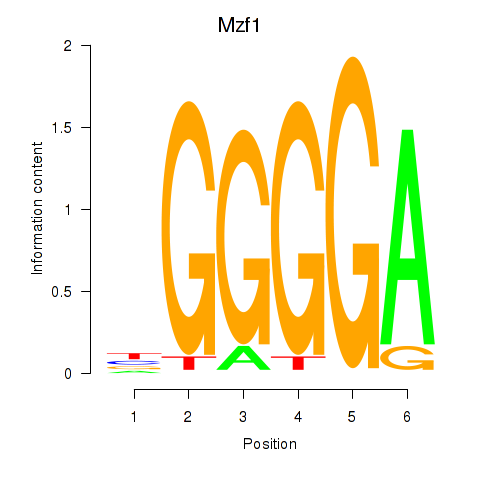
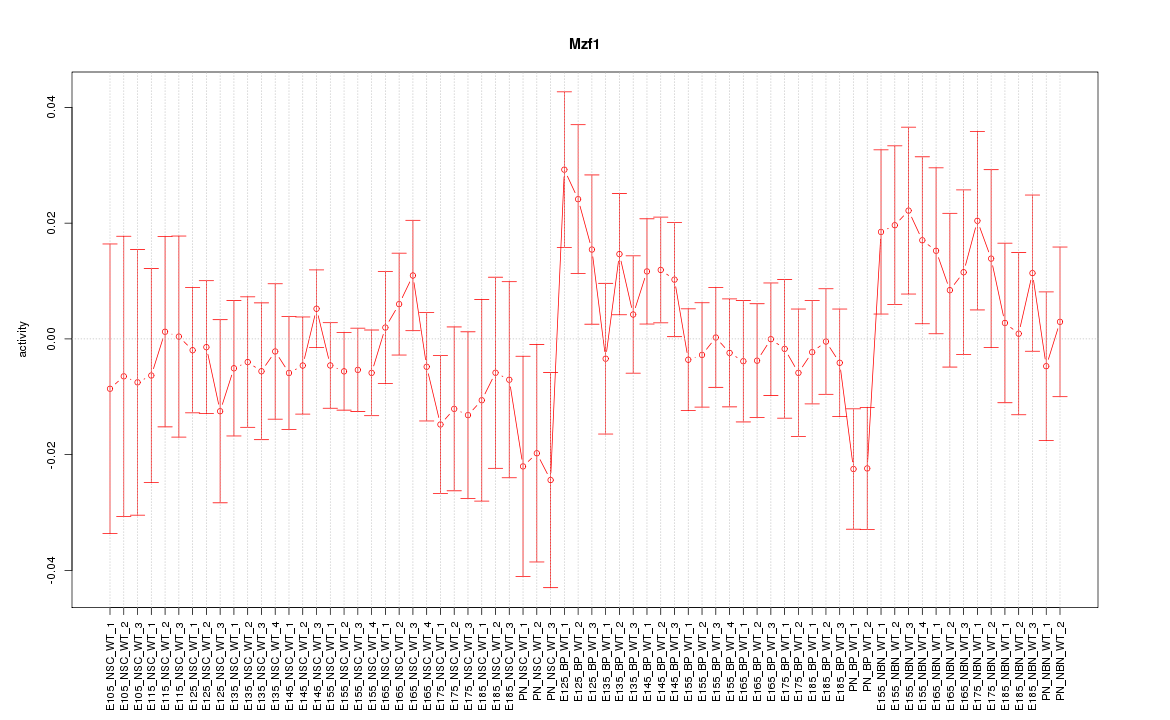
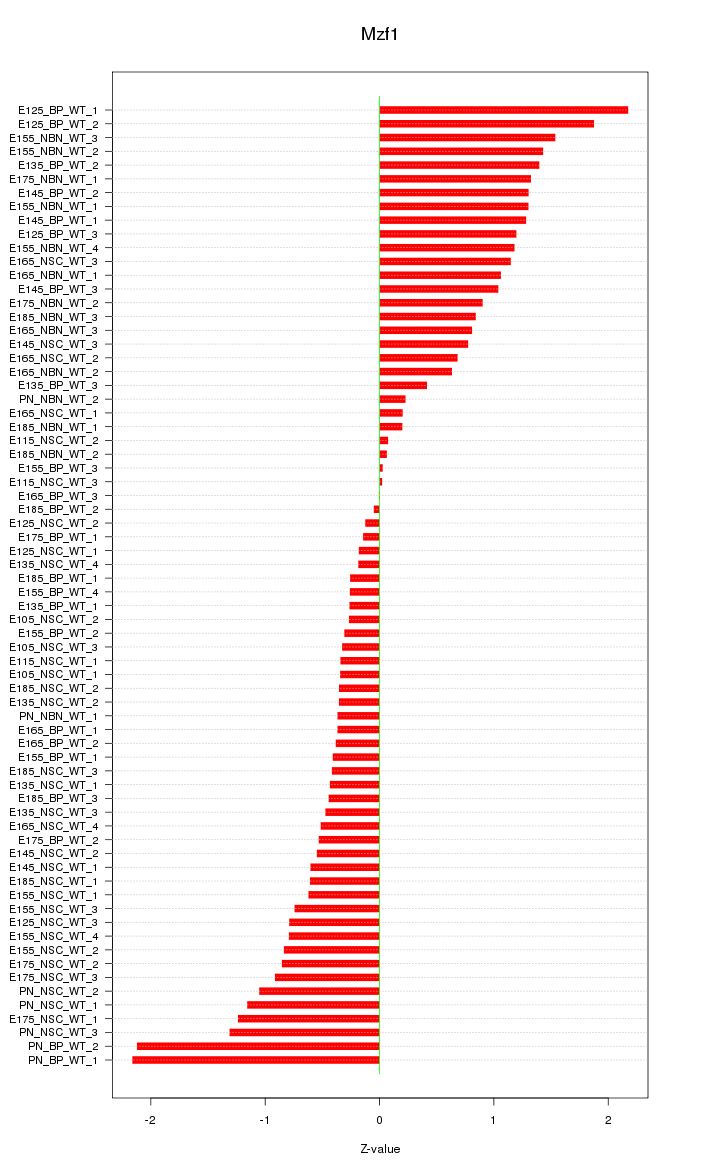
| 2.1 |
6.3 |
GO:0060478 |
acrosomal vesicle exocytosis(GO:0060478) |
| 1.9 |
5.7 |
GO:0021577 |
hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 1.6 |
4.7 |
GO:0097623 |
potassium ion export across plasma membrane(GO:0097623) |
| 1.6 |
4.7 |
GO:0017055 |
negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.3 |
6.4 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 1.2 |
18.9 |
GO:2000821 |
regulation of grooming behavior(GO:2000821) |
| 1.2 |
1.2 |
GO:0051964 |
negative regulation of synapse assembly(GO:0051964) |
| 1.2 |
3.5 |
GO:0003219 |
cardiac right ventricle formation(GO:0003219) |
| 1.2 |
3.5 |
GO:0021649 |
vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.1 |
3.3 |
GO:2000852 |
regulation of corticosterone secretion(GO:2000852) |
| 0.9 |
2.7 |
GO:0060596 |
mammary placode formation(GO:0060596) |
| 0.9 |
2.7 |
GO:0032430 |
positive regulation of phospholipase A2 activity(GO:0032430) activation of meiosis involved in egg activation(GO:0060466) |
| 0.8 |
3.3 |
GO:1903802 |
L-glutamate(1-) import into cell(GO:1903802) L-glutamate import into cell(GO:1990123) |
| 0.8 |
2.5 |
GO:0033159 |
negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.8 |
2.5 |
GO:0002159 |
desmosome assembly(GO:0002159) |
| 0.8 |
6.6 |
GO:0005513 |
detection of calcium ion(GO:0005513) |
| 0.8 |
1.6 |
GO:0003096 |
renal sodium ion transport(GO:0003096) |
| 0.7 |
4.5 |
GO:0045162 |
clustering of voltage-gated sodium channels(GO:0045162) |
| 0.7 |
2.2 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.7 |
1.5 |
GO:0070212 |
protein poly-ADP-ribosylation(GO:0070212) |
| 0.7 |
3.6 |
GO:0090273 |
regulation of somatostatin secretion(GO:0090273) |
| 0.7 |
2.2 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.7 |
3.5 |
GO:0006680 |
glucosylceramide catabolic process(GO:0006680) |
| 0.7 |
4.0 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.7 |
5.2 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.6 |
5.0 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.6 |
3.1 |
GO:1902683 |
regulation of receptor localization to synapse(GO:1902683) |
| 0.6 |
2.4 |
GO:0060024 |
rhythmic synaptic transmission(GO:0060024) |
| 0.6 |
3.0 |
GO:0031915 |
positive regulation of synaptic plasticity(GO:0031915) |
| 0.6 |
5.3 |
GO:0071420 |
cellular response to histamine(GO:0071420) |
| 0.6 |
1.8 |
GO:1903244 |
positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.6 |
1.2 |
GO:0014735 |
regulation of muscle atrophy(GO:0014735) |
| 0.6 |
2.9 |
GO:0043314 |
negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.6 |
1.7 |
GO:1990034 |
calcium ion export from cell(GO:1990034) |
| 0.6 |
2.3 |
GO:2000304 |
positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.6 |
4.5 |
GO:0035726 |
common myeloid progenitor cell proliferation(GO:0035726) |
| 0.5 |
2.7 |
GO:0002121 |
inter-male aggressive behavior(GO:0002121) |
| 0.5 |
2.2 |
GO:1900122 |
positive regulation of receptor binding(GO:1900122) |
| 0.5 |
1.6 |
GO:0050915 |
sensory perception of sour taste(GO:0050915) |
| 0.5 |
2.7 |
GO:0071638 |
negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.5 |
2.7 |
GO:0001661 |
conditioned taste aversion(GO:0001661) |
| 0.5 |
2.1 |
GO:0051562 |
negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.5 |
1.5 |
GO:0086013 |
membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.5 |
3.5 |
GO:0034047 |
regulation of protein phosphatase type 2A activity(GO:0034047) |
| 0.5 |
7.0 |
GO:0071625 |
vocalization behavior(GO:0071625) |
| 0.5 |
2.5 |
GO:2000587 |
regulation of phospholipase A2 activity(GO:0032429) regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000586) negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.5 |
4.4 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.5 |
1.4 |
GO:1902219 |
regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902218) negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.5 |
1.4 |
GO:0097475 |
motor neuron migration(GO:0097475) spinal cord motor neuron migration(GO:0097476) |
| 0.5 |
2.8 |
GO:0098828 |
modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.5 |
2.8 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.5 |
1.4 |
GO:0030827 |
negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 0.5 |
1.8 |
GO:1904936 |
cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.4 |
3.1 |
GO:0034393 |
positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.4 |
1.3 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.4 |
1.3 |
GO:0035435 |
phosphate ion transmembrane transport(GO:0035435) |
| 0.4 |
8.7 |
GO:1903861 |
regulation of dendrite extension(GO:1903859) positive regulation of dendrite extension(GO:1903861) |
| 0.4 |
1.3 |
GO:0001543 |
ovarian follicle rupture(GO:0001543) |
| 0.4 |
2.0 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.4 |
7.3 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.4 |
1.2 |
GO:1904457 |
positive regulation of neuronal action potential(GO:1904457) |
| 0.4 |
4.0 |
GO:0030322 |
stabilization of membrane potential(GO:0030322) |
| 0.4 |
2.4 |
GO:0032811 |
negative regulation of gamma-aminobutyric acid secretion(GO:0014053) negative regulation of epinephrine secretion(GO:0032811) |
| 0.4 |
1.5 |
GO:0034184 |
positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.4 |
1.1 |
GO:0001919 |
regulation of receptor recycling(GO:0001919) |
| 0.4 |
2.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.4 |
2.5 |
GO:1900194 |
negative regulation of oocyte maturation(GO:1900194) |
| 0.4 |
1.8 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 |
1.4 |
GO:0097118 |
neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 |
2.1 |
GO:0003406 |
retinal pigment epithelium development(GO:0003406) |
| 0.3 |
2.4 |
GO:0007195 |
adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.3 |
1.3 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.3 |
2.0 |
GO:0006177 |
GMP biosynthetic process(GO:0006177) |
| 0.3 |
1.0 |
GO:0060023 |
soft palate development(GO:0060023) |
| 0.3 |
3.5 |
GO:1902018 |
negative regulation of cilium assembly(GO:1902018) |
| 0.3 |
1.9 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.3 |
2.5 |
GO:0000395 |
mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 |
0.9 |
GO:0042271 |
susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.3 |
3.9 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 |
1.5 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.3 |
0.9 |
GO:0038003 |
opioid receptor signaling pathway(GO:0038003) regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.3 |
1.8 |
GO:0046880 |
regulation of follicle-stimulating hormone secretion(GO:0046880) follicle-stimulating hormone secretion(GO:0046884) |
| 0.3 |
3.2 |
GO:0010650 |
positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.3 |
2.5 |
GO:0098907 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.3 |
0.8 |
GO:1904177 |
regulation of adipose tissue development(GO:1904177) |
| 0.3 |
1.7 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.3 |
0.8 |
GO:0006421 |
asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.3 |
1.1 |
GO:0019919 |
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) peptidyl-arginine omega-N-methylation(GO:0035247) histone H4-R3 methylation(GO:0043985) |
| 0.3 |
2.7 |
GO:0001956 |
positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.3 |
2.9 |
GO:0007252 |
I-kappaB phosphorylation(GO:0007252) |
| 0.3 |
1.0 |
GO:0043974 |
histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) |
| 0.3 |
1.8 |
GO:0036072 |
intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 |
2.1 |
GO:2000786 |
positive regulation of autophagosome assembly(GO:2000786) |
| 0.2 |
2.2 |
GO:0061577 |
calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 |
2.7 |
GO:0032482 |
Rab protein signal transduction(GO:0032482) |
| 0.2 |
9.6 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.2 |
0.7 |
GO:0071544 |
diphosphoinositol polyphosphate metabolic process(GO:0071543) diphosphoinositol polyphosphate catabolic process(GO:0071544) |
| 0.2 |
3.6 |
GO:1900273 |
positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.2 |
0.9 |
GO:0060591 |
chondroblast differentiation(GO:0060591) |
| 0.2 |
0.7 |
GO:0070459 |
prolactin secretion(GO:0070459) |
| 0.2 |
1.8 |
GO:0036506 |
maintenance of unfolded protein(GO:0036506) protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 |
2.2 |
GO:0048742 |
regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.9 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.2 |
1.1 |
GO:1905098 |
negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.2 |
0.9 |
GO:2000774 |
positive regulation of cellular senescence(GO:2000774) |
| 0.2 |
2.1 |
GO:0007379 |
segment specification(GO:0007379) |
| 0.2 |
2.6 |
GO:0060081 |
membrane hyperpolarization(GO:0060081) |
| 0.2 |
2.0 |
GO:0031111 |
negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 |
1.4 |
GO:0051798 |
positive regulation of hair follicle development(GO:0051798) |
| 0.2 |
3.3 |
GO:0016486 |
peptide hormone processing(GO:0016486) |
| 0.2 |
1.8 |
GO:0018230 |
peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.2 |
2.1 |
GO:1901250 |
regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.2 |
1.3 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.2 |
0.6 |
GO:0002586 |
positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) detection of peptidoglycan(GO:0032499) |
| 0.2 |
0.8 |
GO:0032055 |
negative regulation of translation in response to stress(GO:0032055) |
| 0.2 |
0.6 |
GO:1900193 |
regulation of oocyte maturation(GO:1900193) |
| 0.2 |
0.4 |
GO:0071550 |
death-inducing signaling complex assembly(GO:0071550) |
| 0.2 |
0.7 |
GO:0002069 |
columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.2 |
0.7 |
GO:0046898 |
response to cycloheximide(GO:0046898) |
| 0.2 |
0.5 |
GO:0021698 |
cerebellar cortex structural organization(GO:0021698) |
| 0.2 |
0.5 |
GO:0070172 |
oculomotor nerve development(GO:0021557) positive regulation of tooth mineralization(GO:0070172) |
| 0.2 |
1.5 |
GO:2000601 |
positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 |
2.1 |
GO:0043252 |
oligopeptide transport(GO:0006857) sodium-independent organic anion transport(GO:0043252) |
| 0.2 |
1.9 |
GO:0048672 |
positive regulation of collateral sprouting(GO:0048672) |
| 0.2 |
1.1 |
GO:0071635 |
negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.2 |
3.0 |
GO:0045063 |
T-helper 1 cell differentiation(GO:0045063) |
| 0.2 |
9.8 |
GO:0007019 |
microtubule depolymerization(GO:0007019) |
| 0.2 |
0.9 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.2 |
1.9 |
GO:0043517 |
positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.2 |
0.9 |
GO:0090267 |
positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 |
4.0 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.2 |
0.5 |
GO:0007023 |
post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.2 |
1.1 |
GO:0045742 |
positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.1 |
0.3 |
GO:0034351 |
negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 |
0.4 |
GO:0048162 |
preantral ovarian follicle growth(GO:0001546) multi-layer follicle stage(GO:0048162) |
| 0.1 |
1.0 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.1 |
0.7 |
GO:0021993 |
initiation of neural tube closure(GO:0021993) |
| 0.1 |
1.4 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
0.1 |
GO:0050713 |
negative regulation of interleukin-1 secretion(GO:0050711) negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.1 |
0.5 |
GO:0035624 |
receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 |
4.3 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
0.5 |
GO:0045925 |
positive regulation of female receptivity(GO:0045925) |
| 0.1 |
9.8 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |
| 0.1 |
0.1 |
GO:1901097 |
negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 |
1.2 |
GO:0045906 |
negative regulation of vasoconstriction(GO:0045906) |
| 0.1 |
1.3 |
GO:1902259 |
regulation of delayed rectifier potassium channel activity(GO:1902259) |
| 0.1 |
0.6 |
GO:0023035 |
CD40 signaling pathway(GO:0023035) |
| 0.1 |
0.6 |
GO:0002182 |
cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 |
1.4 |
GO:0048172 |
regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 |
0.4 |
GO:0035928 |
rRNA import into mitochondrion(GO:0035928) |
| 0.1 |
1.6 |
GO:0048280 |
vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.1 |
2.1 |
GO:1902857 |
positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.1 |
1.8 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.1 |
1.2 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.4 |
GO:0086046 |
membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.1 |
2.7 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.1 |
2.6 |
GO:0043171 |
peptide catabolic process(GO:0043171) |
| 0.1 |
1.0 |
GO:0035280 |
miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.1 |
0.3 |
GO:0032241 |
positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 |
1.3 |
GO:0051601 |
exocyst localization(GO:0051601) |
| 0.1 |
0.5 |
GO:0030576 |
Cajal body organization(GO:0030576) |
| 0.1 |
0.5 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 |
0.6 |
GO:0015671 |
oxygen transport(GO:0015671) |
| 0.1 |
3.3 |
GO:0016578 |
histone deubiquitination(GO:0016578) |
| 0.1 |
0.6 |
GO:0090666 |
scaRNA localization to Cajal body(GO:0090666) |
| 0.1 |
2.8 |
GO:0061098 |
positive regulation of protein tyrosine kinase activity(GO:0061098) |
| 0.1 |
0.3 |
GO:2000501 |
natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.1 |
1.5 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 |
0.3 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 |
0.9 |
GO:0007288 |
sperm axoneme assembly(GO:0007288) protein polyglutamylation(GO:0018095) |
| 0.1 |
0.5 |
GO:0051189 |
Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.1 |
2.4 |
GO:0034063 |
stress granule assembly(GO:0034063) |
| 0.1 |
0.5 |
GO:0051043 |
regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 |
0.3 |
GO:0060334 |
regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.1 |
0.6 |
GO:0044375 |
regulation of peroxisome size(GO:0044375) |
| 0.1 |
0.9 |
GO:0090527 |
actin filament reorganization(GO:0090527) |
| 0.1 |
0.4 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.1 |
0.6 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.2 |
GO:2000551 |
regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.1 |
1.5 |
GO:0046069 |
cGMP catabolic process(GO:0046069) |
| 0.1 |
4.3 |
GO:0046513 |
ceramide biosynthetic process(GO:0046513) |
| 0.1 |
0.5 |
GO:0030263 |
apoptotic chromosome condensation(GO:0030263) |
| 0.1 |
1.1 |
GO:0009072 |
aromatic amino acid family metabolic process(GO:0009072) |
| 0.1 |
1.5 |
GO:0048305 |
immunoglobulin secretion(GO:0048305) |
| 0.1 |
0.2 |
GO:0044339 |
canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 |
0.8 |
GO:0010839 |
negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 |
0.2 |
GO:1903378 |
positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.1 |
0.5 |
GO:0071243 |
cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 |
0.2 |
GO:0030828 |
positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.1 |
0.2 |
GO:0009446 |
putrescine biosynthetic process(GO:0009446) |
| 0.1 |
0.8 |
GO:0021942 |
radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 |
0.2 |
GO:1901187 |
regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 |
0.6 |
GO:1904707 |
positive regulation of vascular smooth muscle cell proliferation(GO:1904707) |
| 0.1 |
0.7 |
GO:0019800 |
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 |
1.4 |
GO:0048741 |
skeletal muscle fiber development(GO:0048741) |
| 0.1 |
0.4 |
GO:0098734 |
protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 |
0.7 |
GO:0051968 |
positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.1 |
1.1 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.1 |
0.9 |
GO:2001241 |
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.1 |
0.6 |
GO:0043084 |
penile erection(GO:0043084) |
| 0.1 |
0.4 |
GO:0019695 |
choline metabolic process(GO:0019695) neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 |
0.2 |
GO:0018343 |
protein farnesylation(GO:0018343) |
| 0.1 |
0.4 |
GO:1902309 |
negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 |
0.3 |
GO:0097501 |
stress response to metal ion(GO:0097501) |
| 0.1 |
0.3 |
GO:0043567 |
regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.1 |
0.2 |
GO:0072531 |
pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.1 |
1.3 |
GO:0035024 |
negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 |
0.3 |
GO:0002710 |
negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 |
0.2 |
GO:0001827 |
inner cell mass cell fate commitment(GO:0001827) |
| 0.1 |
0.3 |
GO:2000676 |
positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.1 |
0.3 |
GO:0072592 |
oxygen metabolic process(GO:0072592) |
| 0.1 |
1.1 |
GO:1901381 |
positive regulation of potassium ion transmembrane transport(GO:1901381) |
| 0.1 |
3.4 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 |
1.0 |
GO:0080182 |
histone H3-K4 trimethylation(GO:0080182) |
| 0.1 |
0.4 |
GO:2001223 |
negative regulation of neuron migration(GO:2001223) |
| 0.1 |
1.3 |
GO:0031110 |
regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 |
0.1 |
GO:2000393 |
negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 |
0.9 |
GO:0045793 |
positive regulation of cell size(GO:0045793) |
| 0.1 |
2.2 |
GO:0006110 |
regulation of glycolytic process(GO:0006110) |
| 0.1 |
0.5 |
GO:0060013 |
righting reflex(GO:0060013) positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 |
0.2 |
GO:0072675 |
osteoclast fusion(GO:0072675) |
| 0.1 |
0.4 |
GO:0030854 |
positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 |
0.4 |
GO:0048702 |
embryonic neurocranium morphogenesis(GO:0048702) |
| 0.1 |
0.2 |
GO:0010459 |
negative regulation of heart rate(GO:0010459) |
| 0.1 |
3.6 |
GO:0001578 |
microtubule bundle formation(GO:0001578) |
| 0.1 |
0.9 |
GO:0043984 |
histone H4-K16 acetylation(GO:0043984) |
| 0.1 |
0.5 |
GO:0075522 |
IRES-dependent viral translational initiation(GO:0075522) |
| 0.1 |
0.3 |
GO:0043569 |
negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 |
0.6 |
GO:0070884 |
regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.1 |
0.2 |
GO:0002922 |
positive regulation of humoral immune response(GO:0002922) |
| 0.1 |
0.3 |
GO:1900095 |
regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.1 |
0.8 |
GO:0045717 |
negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.1 |
0.9 |
GO:0051966 |
regulation of synaptic transmission, glutamatergic(GO:0051966) |
| 0.0 |
1.8 |
GO:0046579 |
positive regulation of Ras protein signal transduction(GO:0046579) |
| 0.0 |
0.7 |
GO:0008535 |
respiratory chain complex IV assembly(GO:0008535) |
| 0.0 |
0.5 |
GO:1901841 |
regulation of high voltage-gated calcium channel activity(GO:1901841) |
| 0.0 |
0.5 |
GO:1901385 |
regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 |
0.6 |
GO:0000042 |
protein targeting to Golgi(GO:0000042) |
| 0.0 |
1.1 |
GO:0051865 |
protein autoubiquitination(GO:0051865) |
| 0.0 |
0.2 |
GO:0060368 |
regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 |
0.7 |
GO:0060218 |
hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 |
1.2 |
GO:0046580 |
negative regulation of Ras protein signal transduction(GO:0046580) |
| 0.0 |
0.6 |
GO:0043508 |
negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 |
0.1 |
GO:0061428 |
negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 |
0.2 |
GO:0007608 |
sensory perception of smell(GO:0007608) |
| 0.0 |
0.7 |
GO:0031167 |
rRNA methylation(GO:0031167) |
| 0.0 |
0.8 |
GO:0060384 |
innervation(GO:0060384) |
| 0.0 |
0.3 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.0 |
2.4 |
GO:0070936 |
protein K48-linked ubiquitination(GO:0070936) |
| 0.0 |
0.2 |
GO:0001831 |
trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 |
0.8 |
GO:0010971 |
positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 |
0.3 |
GO:0002643 |
regulation of tolerance induction(GO:0002643) |
| 0.0 |
0.6 |
GO:0032927 |
positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 |
0.2 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.0 |
1.4 |
GO:0030010 |
establishment of cell polarity(GO:0030010) |
| 0.0 |
0.6 |
GO:0045773 |
positive regulation of axon extension(GO:0045773) |
| 0.0 |
0.6 |
GO:0045116 |
protein neddylation(GO:0045116) |
| 0.0 |
0.2 |
GO:0034227 |
tRNA thio-modification(GO:0034227) |
| 0.0 |
0.2 |
GO:0034058 |
endosomal vesicle fusion(GO:0034058) |
| 0.0 |
0.4 |
GO:0045109 |
intermediate filament organization(GO:0045109) |
| 0.0 |
0.1 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 |
0.6 |
GO:0032292 |
myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 |
1.0 |
GO:0031122 |
cytoplasmic microtubule organization(GO:0031122) |
| 0.0 |
0.8 |
GO:0006625 |
protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 |
1.1 |
GO:0007029 |
endoplasmic reticulum organization(GO:0007029) |
| 0.0 |
0.2 |
GO:1904690 |
regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 |
0.8 |
GO:0071806 |
intracellular protein transmembrane transport(GO:0065002) protein transmembrane transport(GO:0071806) |
| 0.0 |
0.3 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 |
0.4 |
GO:0006465 |
signal peptide processing(GO:0006465) |
| 0.0 |
0.3 |
GO:0034122 |
negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 |
0.2 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
0.5 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.0 |
0.1 |
GO:0048014 |
Tie signaling pathway(GO:0048014) |
| 0.0 |
0.8 |
GO:0006879 |
cellular iron ion homeostasis(GO:0006879) |
| 0.0 |
1.5 |
GO:0010923 |
negative regulation of phosphatase activity(GO:0010923) |
| 0.0 |
0.0 |
GO:0072367 |
regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 |
0.3 |
GO:0032785 |
negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 0.0 |
1.4 |
GO:0002028 |
regulation of sodium ion transport(GO:0002028) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.5 |
GO:2000785 |
regulation of autophagosome assembly(GO:2000785) |
| 0.0 |
0.1 |
GO:0006624 |
vacuolar protein processing(GO:0006624) |
| 0.0 |
0.2 |
GO:0030252 |
growth hormone secretion(GO:0030252) |
| 0.0 |
1.9 |
GO:0000209 |
protein polyubiquitination(GO:0000209) |
| 0.0 |
0.2 |
GO:0046007 |
negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 |
0.1 |
GO:0061502 |
early endosome to recycling endosome transport(GO:0061502) |
| 0.0 |
2.9 |
GO:0042787 |
protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 |
0.2 |
GO:1900452 |
regulation of long term synaptic depression(GO:1900452) |
| 0.0 |
0.2 |
GO:0000154 |
rRNA modification(GO:0000154) |
| 0.0 |
0.1 |
GO:0090141 |
positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 |
0.5 |
GO:0006584 |
catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.0 |
0.3 |
GO:0080111 |
DNA demethylation(GO:0080111) |
| 0.0 |
0.3 |
GO:0070200 |
establishment of protein localization to telomere(GO:0070200) |
| 0.0 |
4.1 |
GO:0051260 |
protein homooligomerization(GO:0051260) |
| 0.0 |
0.9 |
GO:1903363 |
negative regulation of cellular protein catabolic process(GO:1903363) |
| 0.0 |
0.1 |
GO:0043653 |
mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 |
0.2 |
GO:0048147 |
negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 |
0.3 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.0 |
0.1 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 |
0.3 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 |
0.5 |
GO:0043967 |
histone H4 acetylation(GO:0043967) |
| 0.0 |
0.3 |
GO:0016239 |
positive regulation of macroautophagy(GO:0016239) |
| 0.0 |
0.4 |
GO:0010507 |
negative regulation of autophagy(GO:0010507) |
| 0.0 |
0.1 |
GO:0009404 |
toxin metabolic process(GO:0009404) |
| 0.0 |
0.3 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.2 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.0 |
0.3 |
GO:0032467 |
positive regulation of cytokinesis(GO:0032467) |
| 0.0 |
0.1 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.0 |
GO:0097274 |
urea homeostasis(GO:0097274) |
| 0.0 |
0.5 |
GO:0000184 |
nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 |
0.2 |
GO:0014741 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 |
0.3 |
GO:0043687 |
post-translational protein modification(GO:0043687) |
| 0.0 |
0.1 |
GO:0001731 |
formation of translation preinitiation complex(GO:0001731) |