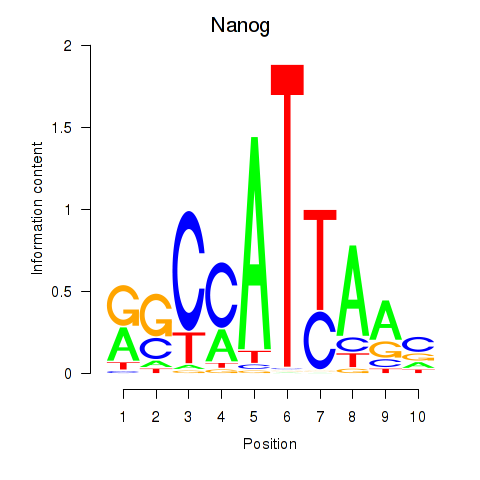
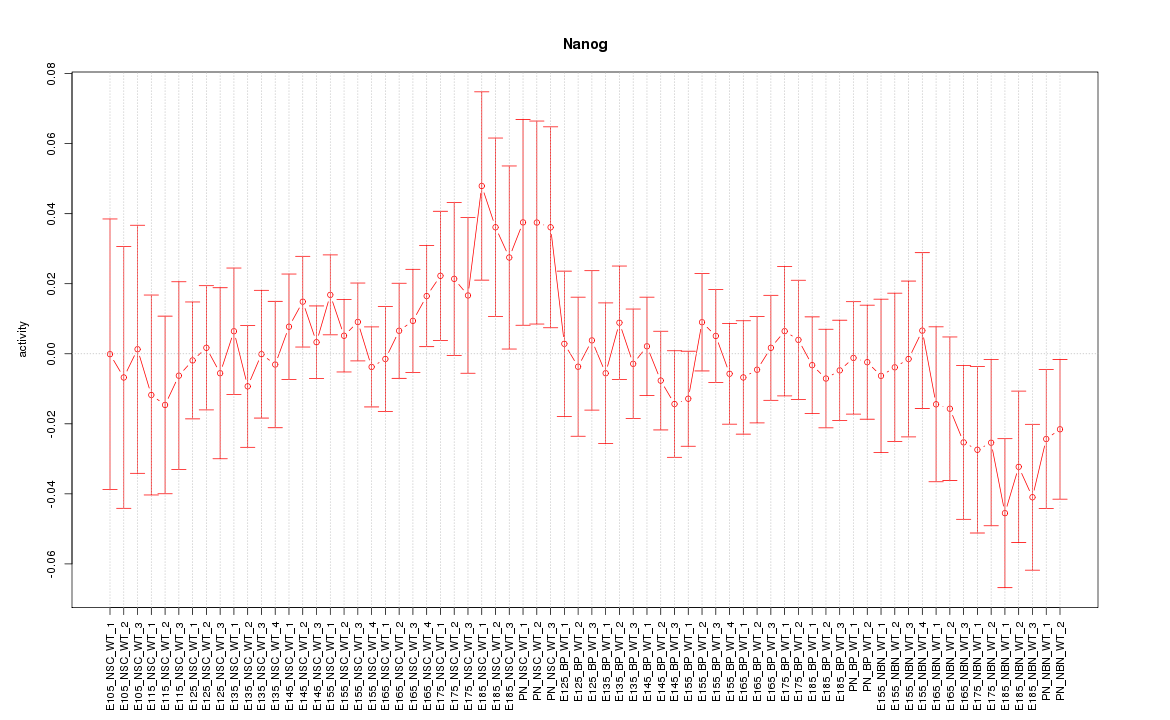
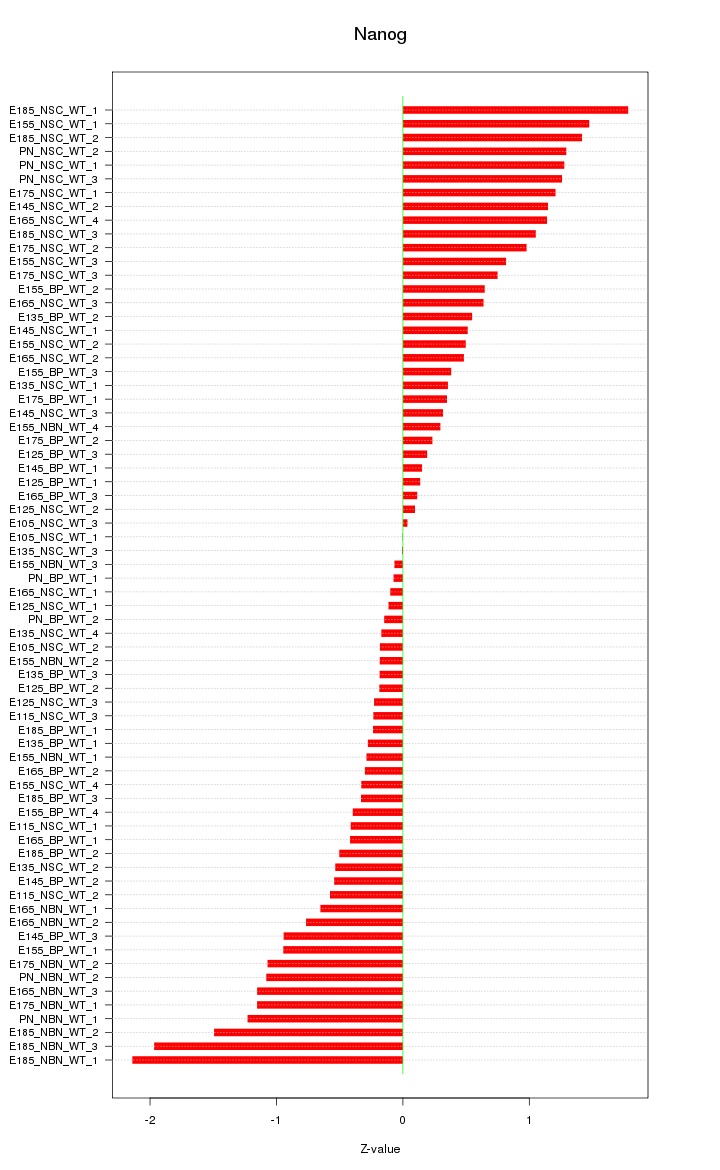
| 3.9 |
15.7 |
GO:2000974 |
negative regulation of pro-B cell differentiation(GO:2000974) |
| 2.6 |
10.2 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 1.8 |
11.0 |
GO:1902847 |
macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 1.7 |
5.1 |
GO:0072092 |
ureteric bud invasion(GO:0072092) |
| 1.1 |
8.8 |
GO:0033601 |
positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 1.1 |
5.3 |
GO:0021764 |
amygdala development(GO:0021764) |
| 1.0 |
7.2 |
GO:0015840 |
urea transport(GO:0015840) urea transmembrane transport(GO:0071918) |
| 1.0 |
4.0 |
GO:0042360 |
vitamin E metabolic process(GO:0042360) |
| 1.0 |
6.8 |
GO:0021780 |
oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.9 |
2.7 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.9 |
14.8 |
GO:0022027 |
interkinetic nuclear migration(GO:0022027) |
| 0.8 |
4.0 |
GO:0014719 |
skeletal muscle satellite cell activation(GO:0014719) |
| 0.7 |
2.0 |
GO:0006553 |
lysine metabolic process(GO:0006553) |
| 0.7 |
8.1 |
GO:0019985 |
translesion synthesis(GO:0019985) |
| 0.7 |
2.0 |
GO:0030862 |
neuroblast division in subventricular zone(GO:0021849) regulation of polarized epithelial cell differentiation(GO:0030860) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.6 |
2.5 |
GO:1904565 |
response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.6 |
2.3 |
GO:0006742 |
NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.6 |
2.3 |
GO:2001032 |
regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.6 |
1.7 |
GO:0019676 |
ammonia assimilation cycle(GO:0019676) |
| 0.4 |
2.7 |
GO:1902261 |
positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.4 |
3.3 |
GO:0033314 |
mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 |
1.5 |
GO:1900220 |
semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.4 |
1.1 |
GO:0061300 |
cerebellum vasculature development(GO:0061300) |
| 0.4 |
1.1 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.4 |
4.0 |
GO:0014883 |
transition between fast and slow fiber(GO:0014883) positive regulation of stem cell population maintenance(GO:1902459) |
| 0.4 |
1.8 |
GO:1904721 |
regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) negative regulation of immunoglobulin secretion(GO:0051025) negative regulation of ribonuclease activity(GO:0060701) negative regulation of endoribonuclease activity(GO:0060702) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.3 |
2.4 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.3 |
0.7 |
GO:0021913 |
regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.3 |
1.0 |
GO:0070671 |
monocyte extravasation(GO:0035696) activation of meiosis involved in egg activation(GO:0060466) response to interleukin-12(GO:0070671) positive regulation of acrosome reaction(GO:2000344) regulation of monocyte extravasation(GO:2000437) |
| 0.3 |
2.4 |
GO:1900262 |
regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 |
1.4 |
GO:0042997 |
negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.3 |
3.9 |
GO:0071803 |
positive regulation of podosome assembly(GO:0071803) |
| 0.3 |
1.1 |
GO:0071139 |
resolution of recombination intermediates(GO:0071139) |
| 0.3 |
2.0 |
GO:0071361 |
cellular response to ethanol(GO:0071361) |
| 0.3 |
1.1 |
GO:0030647 |
polyketide metabolic process(GO:0030638) aminoglycoside antibiotic metabolic process(GO:0030647) daunorubicin metabolic process(GO:0044597) doxorubicin metabolic process(GO:0044598) |
| 0.3 |
3.5 |
GO:0048706 |
embryonic skeletal system development(GO:0048706) |
| 0.3 |
1.9 |
GO:0030579 |
ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.3 |
1.0 |
GO:0018242 |
protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 |
1.0 |
GO:0071596 |
ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 |
1.9 |
GO:0044027 |
hypermethylation of CpG island(GO:0044027) |
| 0.2 |
2.2 |
GO:0034983 |
peptidyl-lysine deacetylation(GO:0034983) regulation of skeletal muscle fiber development(GO:0048742) |
| 0.2 |
0.6 |
GO:2000642 |
negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 |
1.3 |
GO:1901029 |
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 |
1.6 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
3.5 |
GO:0007530 |
sex determination(GO:0007530) |
| 0.2 |
1.3 |
GO:0071732 |
cellular response to nitric oxide(GO:0071732) |
| 0.2 |
0.5 |
GO:0002014 |
vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.2 |
1.2 |
GO:0006361 |
transcription initiation from RNA polymerase I promoter(GO:0006361) termination of RNA polymerase I transcription(GO:0006363) |
| 0.2 |
0.5 |
GO:0010986 |
complement receptor mediated signaling pathway(GO:0002430) GPI anchor release(GO:0006507) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.2 |
1.0 |
GO:0035989 |
tendon development(GO:0035989) |
| 0.2 |
2.6 |
GO:0071168 |
protein localization to chromatin(GO:0071168) |
| 0.2 |
0.8 |
GO:0010991 |
negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.6 |
GO:0035521 |
monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 |
2.9 |
GO:0001886 |
endothelial cell morphogenesis(GO:0001886) |
| 0.1 |
0.8 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 |
1.1 |
GO:0018065 |
protein-cofactor linkage(GO:0018065) |
| 0.1 |
1.4 |
GO:0000185 |
activation of MAPKKK activity(GO:0000185) |
| 0.1 |
0.7 |
GO:0007217 |
tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 |
2.8 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
0.8 |
GO:2000301 |
negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 |
0.5 |
GO:0010616 |
negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of protein glycosylation(GO:0060051) |
| 0.1 |
3.0 |
GO:0034508 |
centromere complex assembly(GO:0034508) |
| 0.1 |
5.7 |
GO:0005978 |
glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.1 |
2.3 |
GO:1902043 |
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 |
0.3 |
GO:0019065 |
receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 |
2.6 |
GO:0034723 |
DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 |
0.2 |
GO:1903895 |
negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.1 |
3.8 |
GO:0071300 |
cellular response to retinoic acid(GO:0071300) |
| 0.1 |
1.2 |
GO:0050860 |
negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.1 |
7.1 |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.1 |
3.9 |
GO:0043666 |
regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.1 |
0.5 |
GO:0089711 |
L-glutamate transmembrane transport(GO:0089711) |
| 0.1 |
4.7 |
GO:0051489 |
regulation of filopodium assembly(GO:0051489) |
| 0.1 |
0.9 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.1 |
1.0 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.9 |
GO:0051451 |
myoblast migration(GO:0051451) |
| 0.1 |
0.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 |
0.8 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
2.5 |
GO:0007212 |
dopamine receptor signaling pathway(GO:0007212) |
| 0.1 |
0.9 |
GO:0006388 |
tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 |
2.2 |
GO:0031572 |
G2 DNA damage checkpoint(GO:0031572) |
| 0.1 |
1.2 |
GO:0045907 |
positive regulation of vasoconstriction(GO:0045907) |
| 0.1 |
1.7 |
GO:0036075 |
endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.1 |
0.2 |
GO:0060489 |
epicardial cell to mesenchymal cell transition(GO:0003347) establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.1 |
1.8 |
GO:0050873 |
brown fat cell differentiation(GO:0050873) |
| 0.1 |
0.5 |
GO:0051131 |
chaperone-mediated protein complex assembly(GO:0051131) |
| 0.1 |
0.2 |
GO:1903298 |
regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) |
| 0.1 |
0.5 |
GO:0007202 |
activation of phospholipase C activity(GO:0007202) |
| 0.0 |
0.1 |
GO:0030070 |
insulin processing(GO:0030070) |
| 0.0 |
1.1 |
GO:0033539 |
fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 |
0.5 |
GO:0016127 |
cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 |
0.5 |
GO:2001022 |
positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.0 |
1.9 |
GO:0003341 |
cilium movement(GO:0003341) |
| 0.0 |
0.4 |
GO:0034242 |
negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 |
0.3 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.0 |
0.2 |
GO:2001268 |
positive regulation of synaptic plasticity(GO:0031915) positive regulation of receptor binding(GO:1900122) negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.0 |
1.3 |
GO:0006376 |
mRNA splice site selection(GO:0006376) |
| 0.0 |
0.4 |
GO:0097186 |
amelogenesis(GO:0097186) |
| 0.0 |
0.7 |
GO:0006298 |
mismatch repair(GO:0006298) |
| 0.0 |
1.1 |
GO:0045744 |
negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 |
0.6 |
GO:2000058 |
regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 |
1.9 |
GO:0030148 |
sphingolipid biosynthetic process(GO:0030148) |
| 0.0 |
1.5 |
GO:0007173 |
epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 |
0.2 |
GO:0016322 |
neuron remodeling(GO:0016322) |
| 0.0 |
3.1 |
GO:0000910 |
cytokinesis(GO:0000910) |
| 0.0 |
3.5 |
GO:0030833 |
regulation of actin filament polymerization(GO:0030833) |
| 0.0 |
2.2 |
GO:0072332 |
intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 |
0.2 |
GO:0097178 |
ruffle assembly(GO:0097178) |
| 0.0 |
1.5 |
GO:0032092 |
positive regulation of protein binding(GO:0032092) |
| 0.0 |
0.8 |
GO:0002474 |
antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 |
3.8 |
GO:0008654 |
phospholipid biosynthetic process(GO:0008654) |
| 0.0 |
2.7 |
GO:0098780 |
mitophagy in response to mitochondrial depolarization(GO:0098779) response to mitochondrial depolarisation(GO:0098780) |
| 0.0 |
1.2 |
GO:0046785 |
microtubule polymerization(GO:0046785) |
| 0.0 |
0.3 |
GO:0006471 |
protein ADP-ribosylation(GO:0006471) |
| 0.0 |
1.2 |
GO:0051297 |
centrosome organization(GO:0051297) |
| 0.0 |
0.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.0 |
4.2 |
GO:0007067 |
mitotic nuclear division(GO:0007067) |
| 0.0 |
0.4 |
GO:0046329 |
negative regulation of JNK cascade(GO:0046329) |
| 0.0 |
0.6 |
GO:0000380 |
alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 |
1.1 |
GO:0042552 |
myelination(GO:0042552) |
| 0.0 |
0.4 |
GO:0043966 |
histone H3 acetylation(GO:0043966) |