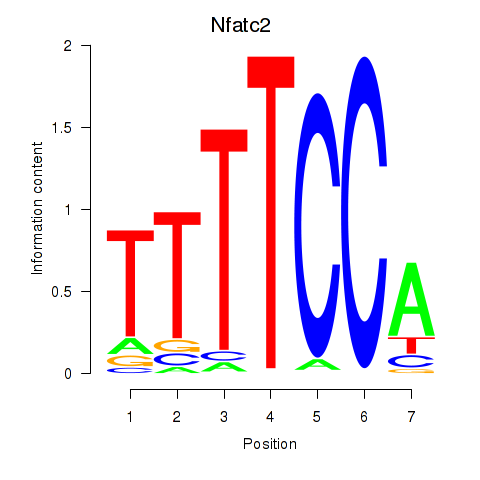
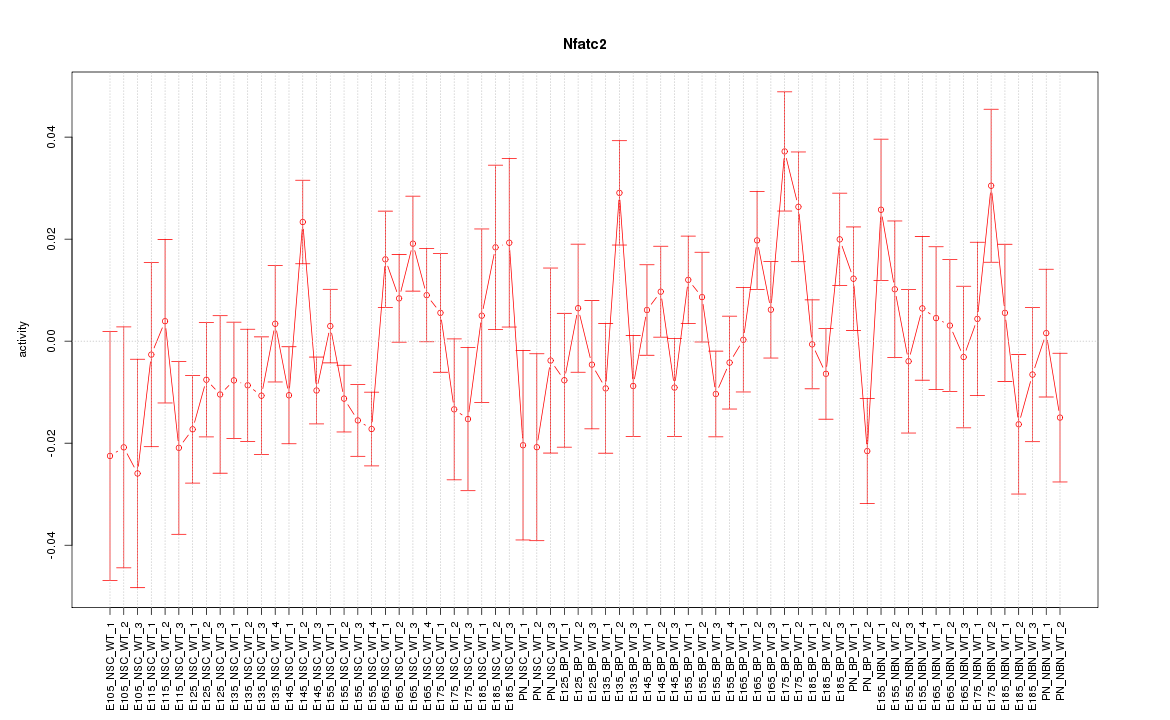
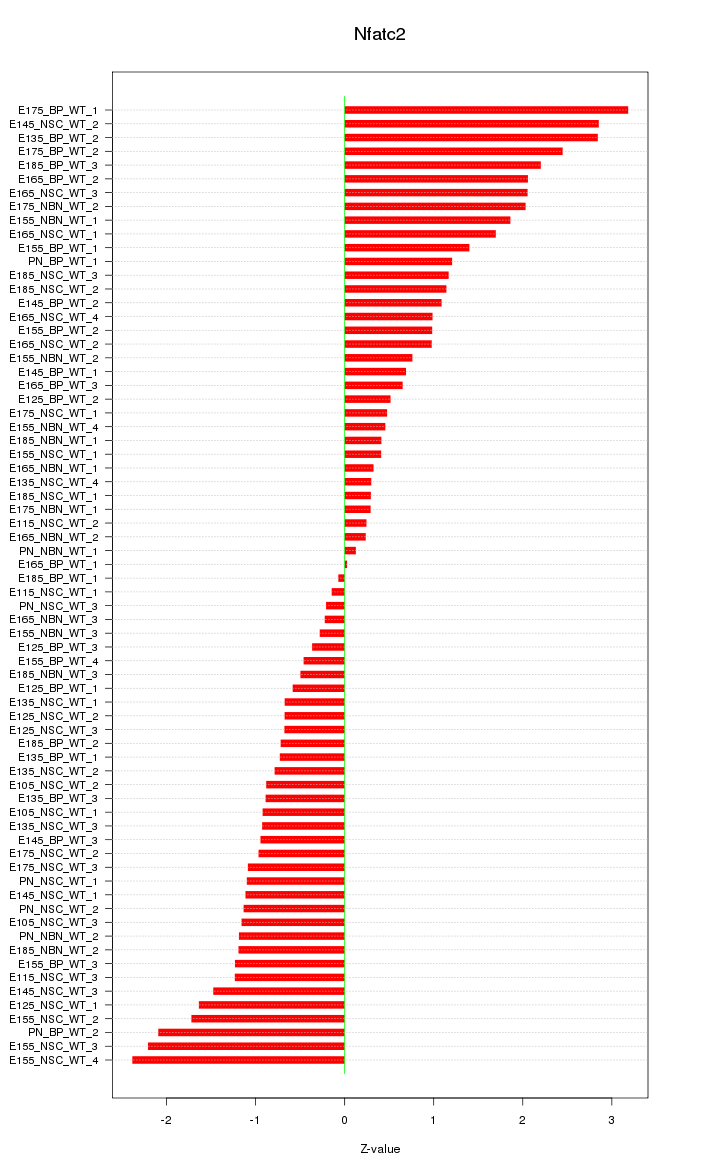
| 2.4 |
9.7 |
GO:0035063 |
nuclear speck organization(GO:0035063) |
| 2.2 |
9.0 |
GO:2000795 |
negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 1.2 |
3.7 |
GO:0060060 |
post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.2 |
5.9 |
GO:0097116 |
gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 1.0 |
5.2 |
GO:1901843 |
positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 1.0 |
2.9 |
GO:0009405 |
pathogenesis(GO:0009405) |
| 0.9 |
15.1 |
GO:0021902 |
commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.9 |
2.7 |
GO:0034334 |
adherens junction maintenance(GO:0034334) |
| 0.8 |
2.5 |
GO:0038095 |
positive regulation of mast cell cytokine production(GO:0032765) Fc-epsilon receptor signaling pathway(GO:0038095) |
| 0.8 |
4.2 |
GO:0098735 |
positive regulation of the force of heart contraction(GO:0098735) |
| 0.8 |
2.5 |
GO:0014877 |
response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.8 |
2.4 |
GO:0031117 |
positive regulation of microtubule depolymerization(GO:0031117) |
| 0.8 |
3.1 |
GO:0021564 |
glossopharyngeal nerve development(GO:0021563) vagus nerve development(GO:0021564) |
| 0.8 |
3.0 |
GO:2000211 |
regulation of glutamate metabolic process(GO:2000211) |
| 0.8 |
2.3 |
GO:0070428 |
regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) |
| 0.7 |
5.2 |
GO:0050908 |
detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.7 |
0.7 |
GO:0048859 |
formation of anatomical boundary(GO:0048859) |
| 0.7 |
4.2 |
GO:1903056 |
regulation of lens fiber cell differentiation(GO:1902746) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.7 |
2.8 |
GO:0021773 |
striatal medium spiny neuron differentiation(GO:0021773) |
| 0.7 |
2.8 |
GO:1903031 |
regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.6 |
2.5 |
GO:0021785 |
branchiomotor neuron axon guidance(GO:0021785) |
| 0.6 |
1.9 |
GO:0044028 |
DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.6 |
3.1 |
GO:0045053 |
protein retention in Golgi apparatus(GO:0045053) |
| 0.6 |
10.9 |
GO:0021860 |
pyramidal neuron development(GO:0021860) |
| 0.6 |
4.2 |
GO:2000809 |
positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.6 |
1.8 |
GO:0038027 |
apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.6 |
1.7 |
GO:1900275 |
negative regulation of phospholipase C activity(GO:1900275) regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.6 |
1.1 |
GO:0090526 |
regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.6 |
4.0 |
GO:0060666 |
dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.6 |
1.7 |
GO:2000111 |
cellular response to parathyroid hormone stimulus(GO:0071374) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.6 |
12.3 |
GO:0035235 |
ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.5 |
1.6 |
GO:1904339 |
negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.5 |
5.3 |
GO:0048149 |
behavioral response to ethanol(GO:0048149) |
| 0.5 |
1.0 |
GO:2000807 |
regulation of synaptic vesicle clustering(GO:2000807) |
| 0.5 |
5.1 |
GO:0042118 |
endothelial cell activation(GO:0042118) |
| 0.5 |
1.5 |
GO:0010871 |
negative regulation of receptor biosynthetic process(GO:0010871) |
| 0.5 |
3.0 |
GO:0000160 |
phosphorelay signal transduction system(GO:0000160) |
| 0.5 |
2.8 |
GO:0072318 |
clathrin coat disassembly(GO:0072318) |
| 0.4 |
4.5 |
GO:0036159 |
inner dynein arm assembly(GO:0036159) |
| 0.4 |
2.2 |
GO:0071476 |
cellular hypotonic response(GO:0071476) |
| 0.4 |
1.3 |
GO:2000620 |
positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.4 |
3.0 |
GO:0043553 |
negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.4 |
2.5 |
GO:0051694 |
pointed-end actin filament capping(GO:0051694) |
| 0.4 |
1.2 |
GO:0002678 |
positive regulation of chronic inflammatory response(GO:0002678) |
| 0.4 |
2.9 |
GO:0042518 |
negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.4 |
2.0 |
GO:0061086 |
negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.4 |
4.0 |
GO:0090315 |
negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 |
1.4 |
GO:0072362 |
regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.3 |
1.7 |
GO:0032224 |
positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.3 |
1.4 |
GO:0072137 |
condensed mesenchymal cell proliferation(GO:0072137) |
| 0.3 |
2.3 |
GO:0090240 |
positive regulation of histone H4 acetylation(GO:0090240) |
| 0.3 |
2.0 |
GO:1903690 |
negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.3 |
1.0 |
GO:0002302 |
CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.3 |
1.3 |
GO:0033326 |
cerebrospinal fluid secretion(GO:0033326) |
| 0.3 |
2.5 |
GO:0061368 |
behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.3 |
2.7 |
GO:0035887 |
aortic smooth muscle cell differentiation(GO:0035887) |
| 0.3 |
2.7 |
GO:0098907 |
protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.3 |
1.5 |
GO:0051013 |
microtubule severing(GO:0051013) |
| 0.3 |
1.5 |
GO:0007256 |
activation of JNKK activity(GO:0007256) |
| 0.3 |
0.6 |
GO:0021814 |
cell motility involved in cerebral cortex radial glia guided migration(GO:0021814) |
| 0.3 |
1.4 |
GO:0010724 |
regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.3 |
1.7 |
GO:1902866 |
regulation of retina development in camera-type eye(GO:1902866) |
| 0.3 |
1.4 |
GO:0071787 |
endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 |
2.4 |
GO:2000467 |
positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 0.3 |
1.6 |
GO:0097368 |
establishment of Sertoli cell barrier(GO:0097368) |
| 0.3 |
1.3 |
GO:0051611 |
negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.3 |
9.5 |
GO:0032728 |
positive regulation of interferon-beta production(GO:0032728) |
| 0.3 |
1.5 |
GO:0003149 |
membranous septum morphogenesis(GO:0003149) |
| 0.2 |
1.5 |
GO:0090383 |
phagosome acidification(GO:0090383) |
| 0.2 |
1.5 |
GO:0071955 |
recycling endosome to Golgi transport(GO:0071955) |
| 0.2 |
1.5 |
GO:0030643 |
cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.2 |
0.7 |
GO:1900369 |
negative regulation of RNA interference(GO:1900369) |
| 0.2 |
1.5 |
GO:0045075 |
interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.2 |
1.9 |
GO:0002674 |
negative regulation of acute inflammatory response(GO:0002674) |
| 0.2 |
3.3 |
GO:0021684 |
cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.2 |
0.7 |
GO:0007228 |
positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.2 |
0.9 |
GO:1901668 |
regulation of superoxide dismutase activity(GO:1901668) |
| 0.2 |
2.8 |
GO:1900383 |
regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 |
0.2 |
GO:0010881 |
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion(GO:0010881) |
| 0.2 |
3.4 |
GO:0031507 |
heterochromatin assembly(GO:0031507) |
| 0.2 |
2.3 |
GO:1900364 |
negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.2 |
0.9 |
GO:1902626 |
assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.2 |
1.7 |
GO:0046541 |
saliva secretion(GO:0046541) |
| 0.2 |
0.6 |
GO:0042097 |
interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 |
1.4 |
GO:1904217 |
regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) regulation of serine C-palmitoyltransferase activity(GO:1904220) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 |
0.8 |
GO:0031642 |
negative regulation of myelination(GO:0031642) |
| 0.2 |
2.2 |
GO:2001256 |
regulation of store-operated calcium entry(GO:2001256) |
| 0.2 |
1.0 |
GO:0018076 |
N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.2 |
4.5 |
GO:0007097 |
nuclear migration(GO:0007097) |
| 0.2 |
1.5 |
GO:0035542 |
regulation of SNARE complex assembly(GO:0035542) |
| 0.2 |
1.5 |
GO:0071557 |
histone H3-K27 demethylation(GO:0071557) |
| 0.2 |
0.8 |
GO:1900017 |
positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.2 |
3.0 |
GO:0002495 |
antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.2 |
2.2 |
GO:0045161 |
neuronal ion channel clustering(GO:0045161) |
| 0.2 |
3.6 |
GO:0071378 |
growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 |
0.5 |
GO:0043323 |
positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.2 |
0.5 |
GO:0003345 |
proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.2 |
3.5 |
GO:0032148 |
activation of protein kinase B activity(GO:0032148) |
| 0.2 |
3.0 |
GO:0007194 |
negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.2 |
0.7 |
GO:0045627 |
positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.2 |
1.2 |
GO:0042984 |
amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 |
0.3 |
GO:1902630 |
regulation of membrane hyperpolarization(GO:1902630) |
| 0.2 |
2.2 |
GO:1901898 |
negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.2 |
0.3 |
GO:0010040 |
response to iron(II) ion(GO:0010040) |
| 0.2 |
7.3 |
GO:0072661 |
protein targeting to plasma membrane(GO:0072661) |
| 0.2 |
2.8 |
GO:2000369 |
regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.2 |
1.2 |
GO:0060267 |
positive regulation of respiratory burst(GO:0060267) |
| 0.2 |
0.5 |
GO:1900365 |
regulation of mRNA polyadenylation(GO:1900363) positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.2 |
1.6 |
GO:0071225 |
cellular response to muramyl dipeptide(GO:0071225) |
| 0.2 |
1.0 |
GO:0019227 |
neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.2 |
0.6 |
GO:0010745 |
negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.2 |
4.5 |
GO:0097320 |
membrane tubulation(GO:0097320) |
| 0.2 |
1.9 |
GO:0000083 |
regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.2 |
3.8 |
GO:0002091 |
negative regulation of receptor internalization(GO:0002091) |
| 0.2 |
0.5 |
GO:0043696 |
dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.2 |
1.3 |
GO:0051797 |
regulation of hair follicle development(GO:0051797) |
| 0.2 |
1.4 |
GO:0009312 |
oligosaccharide biosynthetic process(GO:0009312) |
| 0.2 |
0.8 |
GO:0046684 |
response to pyrethroid(GO:0046684) |
| 0.1 |
0.9 |
GO:0021894 |
cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.1 |
0.4 |
GO:0070649 |
meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 |
2.9 |
GO:0001574 |
ganglioside biosynthetic process(GO:0001574) |
| 0.1 |
0.7 |
GO:0015788 |
UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 |
0.4 |
GO:0017085 |
response to insecticide(GO:0017085) |
| 0.1 |
0.9 |
GO:0097369 |
sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 |
0.6 |
GO:0001920 |
negative regulation of receptor recycling(GO:0001920) |
| 0.1 |
0.9 |
GO:0006488 |
dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 |
0.8 |
GO:0060017 |
parathyroid gland development(GO:0060017) |
| 0.1 |
1.5 |
GO:0033148 |
positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.1 |
2.2 |
GO:0010758 |
regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 |
0.4 |
GO:0048388 |
endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 |
0.7 |
GO:1902897 |
regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 |
1.7 |
GO:2000251 |
positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 |
0.5 |
GO:0060050 |
positive regulation of protein glycosylation(GO:0060050) |
| 0.1 |
1.3 |
GO:0006002 |
fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 |
0.4 |
GO:0019896 |
axonal transport of mitochondrion(GO:0019896) |
| 0.1 |
3.1 |
GO:0032011 |
ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 |
1.2 |
GO:2000651 |
positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 |
3.1 |
GO:0009409 |
response to cold(GO:0009409) |
| 0.1 |
0.4 |
GO:2000643 |
positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.1 |
0.4 |
GO:0035526 |
retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 |
2.0 |
GO:0008090 |
retrograde axonal transport(GO:0008090) |
| 0.1 |
0.9 |
GO:0018026 |
peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 |
0.9 |
GO:0006995 |
cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 |
0.3 |
GO:0097119 |
postsynaptic density protein 95 clustering(GO:0097119) |
| 0.1 |
0.6 |
GO:0035721 |
intraciliary retrograde transport(GO:0035721) |
| 0.1 |
0.9 |
GO:0060693 |
regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.1 |
0.5 |
GO:0035331 |
negative regulation of hippo signaling(GO:0035331) |
| 0.1 |
0.1 |
GO:0072658 |
maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.1 |
0.4 |
GO:0051026 |
chiasma assembly(GO:0051026) |
| 0.1 |
0.6 |
GO:2000270 |
negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.1 |
0.9 |
GO:1903301 |
positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 |
0.9 |
GO:0006622 |
protein targeting to lysosome(GO:0006622) |
| 0.1 |
2.8 |
GO:0048791 |
calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 |
1.3 |
GO:0055059 |
asymmetric neuroblast division(GO:0055059) |
| 0.1 |
1.7 |
GO:0010800 |
positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 |
0.8 |
GO:1901621 |
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 |
0.4 |
GO:0006780 |
uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 |
3.6 |
GO:0061512 |
protein localization to cilium(GO:0061512) |
| 0.1 |
0.6 |
GO:0008635 |
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.1 |
0.3 |
GO:1903977 |
positive regulation of Schwann cell migration(GO:1900149) positive regulation of glial cell migration(GO:1903977) |
| 0.1 |
0.3 |
GO:1900272 |
negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.1 |
0.7 |
GO:0046339 |
diacylglycerol metabolic process(GO:0046339) |
| 0.1 |
0.3 |
GO:2000346 |
negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 |
0.6 |
GO:0021869 |
forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 |
0.6 |
GO:2001054 |
negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 |
1.0 |
GO:0060088 |
auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 |
0.4 |
GO:0070562 |
regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.1 |
0.4 |
GO:0010991 |
regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 |
0.2 |
GO:0042373 |
vitamin K metabolic process(GO:0042373) |
| 0.1 |
1.3 |
GO:0006744 |
ubiquinone biosynthetic process(GO:0006744) |
| 0.1 |
4.5 |
GO:0050775 |
positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 |
1.5 |
GO:0007214 |
gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 |
0.3 |
GO:0033136 |
serine phosphorylation of STAT3 protein(GO:0033136) |
| 0.1 |
0.3 |
GO:0034116 |
positive regulation of heterotypic cell-cell adhesion(GO:0034116) |
| 0.1 |
1.2 |
GO:0005980 |
polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 |
0.5 |
GO:0001880 |
Mullerian duct regression(GO:0001880) osteoblast fate commitment(GO:0002051) |
| 0.1 |
1.9 |
GO:0015991 |
energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.1 |
1.5 |
GO:0007205 |
protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 |
0.8 |
GO:0006883 |
cellular sodium ion homeostasis(GO:0006883) |
| 0.1 |
0.5 |
GO:1901162 |
serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.1 |
0.2 |
GO:0015882 |
L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.1 |
1.0 |
GO:0045792 |
negative regulation of cell size(GO:0045792) |
| 0.1 |
3.0 |
GO:0008542 |
visual learning(GO:0008542) |
| 0.1 |
0.4 |
GO:0030277 |
maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 |
0.6 |
GO:1904424 |
regulation of GTP binding(GO:1904424) |
| 0.1 |
1.0 |
GO:0007271 |
synaptic transmission, cholinergic(GO:0007271) |
| 0.1 |
0.7 |
GO:0019243 |
methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 |
1.0 |
GO:0007130 |
synaptonemal complex assembly(GO:0007130) |
| 0.1 |
0.6 |
GO:0035970 |
peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.1 |
1.3 |
GO:0006415 |
translational termination(GO:0006415) |
| 0.1 |
1.5 |
GO:0071108 |
protein K48-linked deubiquitination(GO:0071108) |
| 0.1 |
0.6 |
GO:0031115 |
negative regulation of microtubule polymerization(GO:0031115) |
| 0.1 |
3.3 |
GO:0033138 |
positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 |
1.6 |
GO:0031571 |
mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.1 |
0.2 |
GO:0000715 |
nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.1 |
0.3 |
GO:1901386 |
negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.1 |
0.2 |
GO:0006436 |
tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 |
1.8 |
GO:0003301 |
physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.1 |
0.5 |
GO:1901223 |
negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 |
0.6 |
GO:0006706 |
steroid catabolic process(GO:0006706) |
| 0.1 |
1.5 |
GO:0018345 |
protein palmitoylation(GO:0018345) |
| 0.1 |
0.3 |
GO:0042780 |
tRNA 3'-end processing(GO:0042780) |
| 0.1 |
1.4 |
GO:0010591 |
regulation of lamellipodium assembly(GO:0010591) |
| 0.1 |
0.9 |
GO:2000311 |
regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 |
0.9 |
GO:0000301 |
retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 |
0.2 |
GO:0044330 |
canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 |
0.7 |
GO:0006903 |
vesicle targeting(GO:0006903) |
| 0.1 |
0.4 |
GO:0015808 |
L-alanine transport(GO:0015808) |
| 0.1 |
1.2 |
GO:0035418 |
protein localization to synapse(GO:0035418) |
| 0.1 |
0.8 |
GO:0036065 |
fucosylation(GO:0036065) |
| 0.1 |
0.3 |
GO:0032308 |
positive regulation of prostaglandin secretion(GO:0032308) |
| 0.1 |
0.2 |
GO:0051708 |
intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) cytosol to ER transport(GO:0046967) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.1 |
0.5 |
GO:0009247 |
glycolipid biosynthetic process(GO:0009247) |
| 0.1 |
0.4 |
GO:0060536 |
cartilage morphogenesis(GO:0060536) |
| 0.1 |
0.7 |
GO:0010614 |
negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.1 |
0.5 |
GO:0043374 |
CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 |
0.4 |
GO:0006013 |
mannose metabolic process(GO:0006013) |
| 0.1 |
1.0 |
GO:0015701 |
bicarbonate transport(GO:0015701) |
| 0.1 |
0.8 |
GO:0031100 |
organ regeneration(GO:0031100) |
| 0.0 |
0.1 |
GO:0006106 |
fumarate metabolic process(GO:0006106) aspartate catabolic process(GO:0006533) alditol biosynthetic process(GO:0019401) |
| 0.0 |
2.0 |
GO:1900006 |
positive regulation of dendrite development(GO:1900006) |
| 0.0 |
0.2 |
GO:1903887 |
motile primary cilium assembly(GO:1903887) |
| 0.0 |
0.7 |
GO:2001258 |
negative regulation of cation channel activity(GO:2001258) |
| 0.0 |
0.5 |
GO:0030157 |
pancreatic juice secretion(GO:0030157) |
| 0.0 |
2.4 |
GO:0015807 |
L-amino acid transport(GO:0015807) |
| 0.0 |
0.4 |
GO:0033564 |
anterior/posterior axon guidance(GO:0033564) |
| 0.0 |
1.4 |
GO:0032091 |
negative regulation of protein binding(GO:0032091) |
| 0.0 |
0.3 |
GO:0032790 |
ribosome disassembly(GO:0032790) |
| 0.0 |
1.0 |
GO:0007638 |
mechanosensory behavior(GO:0007638) |
| 0.0 |
0.5 |
GO:0010669 |
epithelial structure maintenance(GO:0010669) |
| 0.0 |
0.6 |
GO:0010107 |
potassium ion import(GO:0010107) |
| 0.0 |
0.3 |
GO:0035641 |
locomotory exploration behavior(GO:0035641) |
| 0.0 |
0.3 |
GO:0035520 |
monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 |
0.6 |
GO:0032410 |
negative regulation of transporter activity(GO:0032410) |
| 0.0 |
0.6 |
GO:0051382 |
kinetochore assembly(GO:0051382) |
| 0.0 |
0.2 |
GO:0019042 |
viral latency(GO:0019042) |
| 0.0 |
0.1 |
GO:1903896 |
positive regulation of IRE1-mediated unfolded protein response(GO:1903896) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 |
0.9 |
GO:0035329 |
hippo signaling(GO:0035329) |
| 0.0 |
0.1 |
GO:0032929 |
negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 |
0.2 |
GO:0072395 |
signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
| 0.0 |
0.4 |
GO:0090161 |
Golgi ribbon formation(GO:0090161) |
| 0.0 |
0.7 |
GO:0006607 |
NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 |
0.1 |
GO:0006046 |
N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 |
0.1 |
GO:0034316 |
negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 |
0.2 |
GO:0015812 |
gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 |
0.2 |
GO:0042760 |
very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 |
0.6 |
GO:0032456 |
endocytic recycling(GO:0032456) |
| 0.0 |
0.8 |
GO:0031146 |
SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 |
0.3 |
GO:0045579 |
positive regulation of B cell differentiation(GO:0045579) |
| 0.0 |
0.2 |
GO:0061000 |
negative regulation of dendritic spine development(GO:0061000) |
| 0.0 |
0.2 |
GO:0030953 |
astral microtubule organization(GO:0030953) |
| 0.0 |
0.5 |
GO:0045724 |
positive regulation of cilium assembly(GO:0045724) |
| 0.0 |
0.7 |
GO:0000470 |
maturation of LSU-rRNA(GO:0000470) |
| 0.0 |
0.3 |
GO:0035563 |
positive regulation of chromatin binding(GO:0035563) |
| 0.0 |
0.9 |
GO:0045739 |
positive regulation of DNA repair(GO:0045739) |
| 0.0 |
0.3 |
GO:1900745 |
positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 |
0.1 |
GO:0032804 |
negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) |
| 0.0 |
0.1 |
GO:0016338 |
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 |
0.9 |
GO:0043044 |
ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 |
0.4 |
GO:0030520 |
intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 |
0.1 |
GO:0048227 |
plasma membrane to endosome transport(GO:0048227) |
| 0.0 |
0.6 |
GO:0048260 |
positive regulation of receptor-mediated endocytosis(GO:0048260) |
| 0.0 |
0.0 |
GO:0032596 |
protein transport into membrane raft(GO:0032596) |
| 0.0 |
0.6 |
GO:0007218 |
neuropeptide signaling pathway(GO:0007218) |
| 0.0 |
0.2 |
GO:0006654 |
phosphatidic acid biosynthetic process(GO:0006654) phosphatidic acid metabolic process(GO:0046473) |
| 0.0 |
0.7 |
GO:0001658 |
branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 |
0.2 |
GO:0000132 |
establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 |
0.5 |
GO:0051443 |
positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 |
0.9 |
GO:0016575 |
histone deacetylation(GO:0016575) |
| 0.0 |
0.1 |
GO:0045351 |
type I interferon biosynthetic process(GO:0045351) |
| 0.0 |
0.2 |
GO:0002227 |
innate immune response in mucosa(GO:0002227) |
| 0.0 |
0.4 |
GO:0035058 |
nonmotile primary cilium assembly(GO:0035058) |
| 0.0 |
0.1 |
GO:1900748 |
positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 |
0.9 |
GO:0000245 |
spliceosomal complex assembly(GO:0000245) |
| 0.0 |
0.2 |
GO:0016973 |
poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 |
1.1 |
GO:0045814 |
negative regulation of gene expression, epigenetic(GO:0045814) |
| 0.0 |
0.1 |
GO:0044068 |
modulation by symbiont of host cellular process(GO:0044068) |
| 0.0 |
0.2 |
GO:0032755 |
positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 |
0.6 |
GO:0030433 |
ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 |
0.6 |
GO:0031338 |
regulation of vesicle fusion(GO:0031338) |
| 0.0 |
0.1 |
GO:0043267 |
negative regulation of potassium ion transport(GO:0043267) |
| 0.0 |
0.2 |
GO:0048268 |
clathrin coat assembly(GO:0048268) |
| 0.0 |
0.4 |
GO:0045600 |
positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 |
0.5 |
GO:0051965 |
positive regulation of synapse assembly(GO:0051965) |