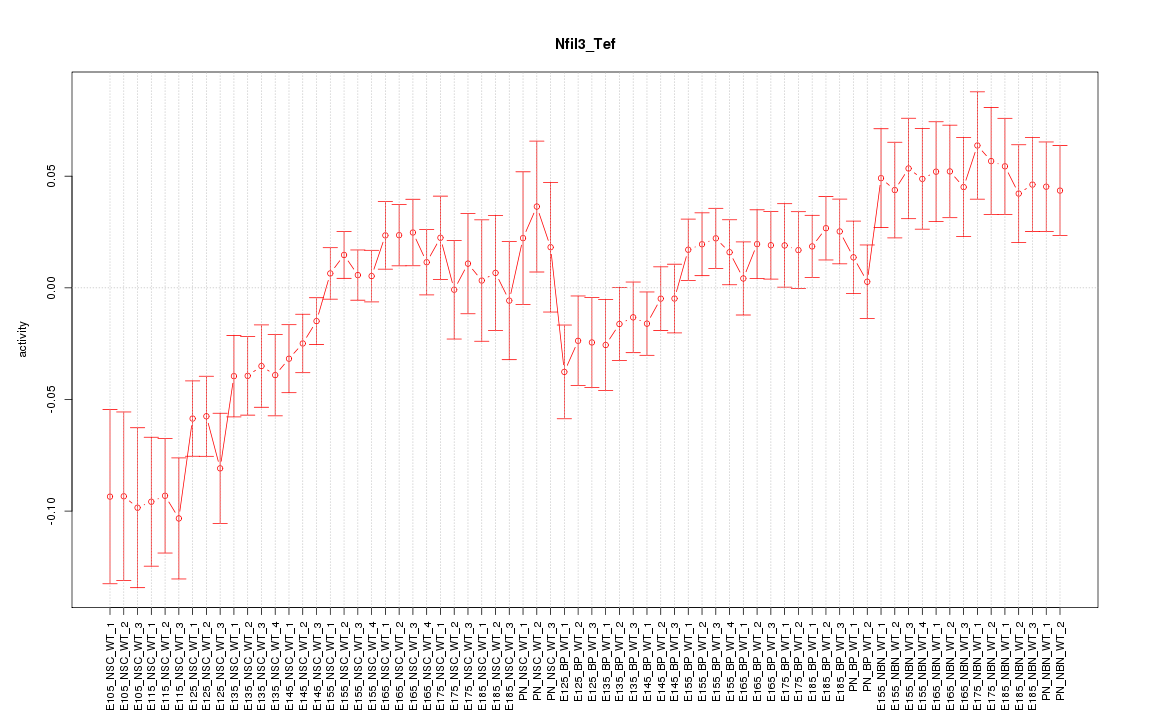
Motif ID: Nfil3_Tef
Z-value: 1.852
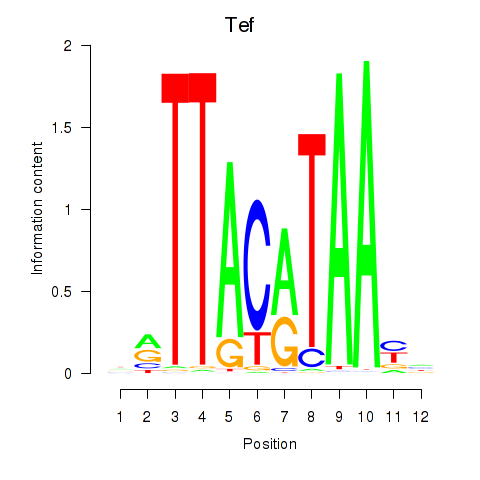
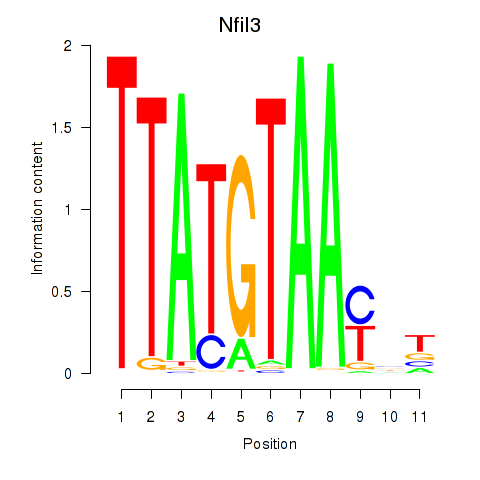
Transcription factors associated with Nfil3_Tef:
| Gene Symbol | Entrez ID | Gene Name |
|---|---|---|
| Nfil3 | ENSMUSG00000056749.7 | Nfil3 |
| Tef | ENSMUSG00000022389.8 | Tef |
Activity-expression correlation:
| Gene Symbol | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tef | mm10_v2_chr15_+_81811414_81811491 | 0.62 | 1.1e-08 | Click! |
| Nfil3 | mm10_v2_chr13_-_52981027_52981083 | 0.57 | 2.4e-07 | Click! |
Top targets:
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 37.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 6.3 | 37.7 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 5.3 | 32.0 | GO:1902847 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) |
| 5.3 | 31.8 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 5.0 | 20.1 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 4.7 | 19.0 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) cellular response to copper ion(GO:0071280) |
| 3.8 | 18.9 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 3.4 | 13.5 | GO:0032289 | central nervous system myelin formation(GO:0032289) cardiac cell fate specification(GO:0060912) |
| 3.0 | 18.2 | GO:0048840 | otolith development(GO:0048840) |
| 3.0 | 12.0 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 2.6 | 7.7 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 2.3 | 6.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) synaptic vesicle lumen acidification(GO:0097401) |
| 2.3 | 45.5 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 2.0 | 10.2 | GO:1900170 | prolactin signaling pathway(GO:0038161) negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 1.9 | 7.8 | GO:0042636 | negative regulation of hair cycle(GO:0042636) negative regulation of hair follicle development(GO:0051799) |
| 1.9 | 5.6 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 1.8 | 7.4 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 1.7 | 15.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.6 | 14.5 | GO:0060373 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 1.5 | 4.4 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 1.3 | 4.0 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 1.3 | 6.3 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 1.3 | 7.5 | GO:0071415 | cellular response to caffeine(GO:0071313) cellular response to purine-containing compound(GO:0071415) |
| 1.2 | 3.7 | GO:1901894 | regulation of calcium-transporting ATPase activity(GO:1901894) |
| 1.2 | 9.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 1.2 | 7.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.1 | 9.0 | GO:0006620 | posttranslational protein targeting to membrane(GO:0006620) |
| 1.1 | 11.1 | GO:0006983 | ER overload response(GO:0006983) |
| 1.1 | 6.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 1.0 | 2.9 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.9 | 6.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.9 | 29.3 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.9 | 11.5 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.9 | 4.4 | GO:1902047 | polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) |
| 0.9 | 7.8 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.8 | 5.5 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.8 | 2.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.8 | 5.4 | GO:0002674 | negative regulation of acute inflammatory response(GO:0002674) |
| 0.8 | 4.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.8 | 8.3 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.7 | 2.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.7 | 1.5 | GO:0002034 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.7 | 3.7 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.7 | 3.6 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.7 | 3.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.7 | 7.0 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.7 | 2.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.6 | 4.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.6 | 3.1 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.6 | 6.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.6 | 1.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.6 | 2.8 | GO:0071476 | cellular hypotonic response(GO:0071476) |
| 0.5 | 1.6 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.5 | 13.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.5 | 3.8 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.5 | 4.8 | GO:0071635 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) negative regulation of transforming growth factor beta production(GO:0071635) |
| 0.5 | 3.6 | GO:0097369 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.5 | 9.6 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 17.6 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.4 | 3.0 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.4 | 3.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.4 | 1.6 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.4 | 2.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.4 | 1.5 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.3 | 4.3 | GO:0036465 | synaptic vesicle recycling(GO:0036465) |
| 0.3 | 1.9 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.3 | 2.6 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.3 | 3.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 2.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 2.9 | GO:0036120 | cellular response to platelet-derived growth factor stimulus(GO:0036120) |
| 0.3 | 2.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.3 | 1.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.2 | 8.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.2 | 0.6 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.2 | 1.4 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 0.5 | GO:0034310 | primary alcohol catabolic process(GO:0034310) |
| 0.2 | 2.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.2 | 2.0 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.2 | 27.6 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.2 | 0.3 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.3 | GO:0045073 | fever generation(GO:0001660) regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) chemokine biosynthetic process(GO:0042033) regulation of chemokine biosynthetic process(GO:0045073) positive regulation of chemokine biosynthetic process(GO:0045080) chemokine metabolic process(GO:0050755) |
| 0.1 | 4.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 2.0 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) |
| 0.1 | 0.4 | GO:2001180 | negative regulation of interleukin-18 production(GO:0032701) negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.1 | 9.9 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.1 | 0.6 | GO:0001575 | globoside metabolic process(GO:0001575) |
| 0.1 | 2.0 | GO:1900037 | regulation of cellular response to hypoxia(GO:1900037) |
| 0.1 | 1.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.1 | 9.5 | GO:0043149 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.1 | 1.7 | GO:0032402 | melanosome transport(GO:0032402) |
| 0.1 | 0.9 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.1 | 0.6 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.1 | 0.5 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.1 | 1.6 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.1 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.1 | 8.1 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 0.3 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.1 | 0.6 | GO:0044539 | long-chain fatty acid import(GO:0044539) |
| 0.1 | 0.1 | GO:0060161 | positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.1 | 5.4 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 11.7 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.1 | 0.3 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.1 | 1.1 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.1 | 0.5 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 2.7 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.1 | 1.8 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 4.0 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 4.1 | GO:0035773 | insulin secretion involved in cellular response to glucose stimulus(GO:0035773) |
| 0.1 | 7.2 | GO:0006836 | neurotransmitter transport(GO:0006836) |
| 0.1 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 1.6 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 1.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0034035 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) |
| 0.0 | 1.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.2 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 1.2 | GO:0055072 | iron ion homeostasis(GO:0055072) |
| 0.0 | 3.3 | GO:0042384 | cilium assembly(GO:0042384) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 32.0 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) neurofibrillary tangle(GO:0097418) |
| 2.6 | 7.8 | GO:0043512 | inhibin A complex(GO:0043512) |
| 1.8 | 28.4 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 1.8 | 19.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 1.6 | 6.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 1.5 | 10.6 | GO:0070695 | FHF complex(GO:0070695) |
| 1.5 | 7.5 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 1.5 | 4.4 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 1.4 | 13.0 | GO:0005883 | neurofilament(GO:0005883) |
| 0.8 | 7.7 | GO:0000805 | X chromosome(GO:0000805) |
| 0.8 | 6.9 | GO:0042581 | specific granule(GO:0042581) |
| 0.6 | 8.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.6 | 11.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 7.4 | GO:0043205 | fibril(GO:0043205) |
| 0.5 | 18.8 | GO:0044298 | cell body membrane(GO:0044298) |
| 0.5 | 35.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.3 | 11.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.3 | 7.9 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 4.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 8.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.3 | 2.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 7.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 3.0 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.2 | 3.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.2 | 25.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 18.9 | GO:0005930 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.2 | 6.4 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 2.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.1 | 3.6 | GO:0005769 | early endosome(GO:0005769) |
| 0.1 | 12.2 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.1 | 0.6 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 7.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 1.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.9 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.1 | 8.8 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.1 | 31.7 | GO:0014069 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.1 | 15.0 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.1 | 2.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.1 | 2.7 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 5.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.1 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 5.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.6 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 4.3 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.1 | 1.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 4.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 3.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 19.5 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 8.1 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 1.6 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 1.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 1.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.6 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 93.2 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 5.1 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.6 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 1.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 3.5 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 4.7 | GO:0000785 | chromatin(GO:0000785) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.3 | 37.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 6.7 | 20.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 3.5 | 10.5 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 3.0 | 12.0 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.7 | 8.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 2.5 | 7.5 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 2.2 | 11.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 2.2 | 6.6 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 2.1 | 14.5 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 1.7 | 6.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 1.7 | 6.8 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 1.6 | 9.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 1.6 | 9.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 1.6 | 6.2 | GO:0031721 | haptoglobin binding(GO:0031720) hemoglobin alpha binding(GO:0031721) |
| 1.4 | 18.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.4 | 32.0 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 1.4 | 8.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 1.3 | 42.0 | GO:0034930 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) heparan sulfate 2-O-sulfotransferase activity(GO:0004394) HNK-1 sulfotransferase activity(GO:0016232) heparan sulfate 6-O-sulfotransferase activity(GO:0017095) trans-9R,10R-dihydrodiolphenanthrene sulfotransferase activity(GO:0018721) 1-phenanthrol sulfotransferase activity(GO:0018722) 3-phenanthrol sulfotransferase activity(GO:0018723) 4-phenanthrol sulfotransferase activity(GO:0018724) trans-3,4-dihydrodiolphenanthrene sulfotransferase activity(GO:0018725) 9-phenanthrol sulfotransferase activity(GO:0018726) 2-phenanthrol sulfotransferase activity(GO:0018727) phenanthrol sulfotransferase activity(GO:0019111) 1-hydroxypyrene sulfotransferase activity(GO:0034930) proteoglycan sulfotransferase activity(GO:0050698) cholesterol sulfotransferase activity(GO:0051922) hydroxyjasmonate sulfotransferase activity(GO:0080131) |
| 1.3 | 7.8 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 1.2 | 4.8 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 1.0 | 15.8 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 1.0 | 2.9 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.9 | 5.4 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.9 | 4.4 | GO:0042979 | ornithine decarboxylase regulator activity(GO:0042979) |
| 0.9 | 14.0 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.8 | 3.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.8 | 2.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.6 | 11.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.6 | 2.9 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.6 | 1.8 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) oncostatin-M receptor activity(GO:0004924) |
| 0.6 | 3.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.5 | 11.5 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.5 | 1.5 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.5 | 2.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.5 | 3.6 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.4 | 7.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.4 | 5.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.4 | 1.6 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.4 | 13.0 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.4 | 7.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.4 | 2.3 | GO:0009374 | biotin binding(GO:0009374) |
| 0.3 | 2.0 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.3 | 2.0 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.3 | 9.1 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.3 | 1.0 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.2 | 2.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.2 | 27.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 4.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.2 | 3.0 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 5.6 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.2 | 0.6 | GO:0043758 | acetate-CoA ligase (ADP-forming) activity(GO:0043758) |
| 0.2 | 1.9 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.2 | 3.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 6.3 | GO:0018602 | sulfonate dioxygenase activity(GO:0000907) 2,4-dichlorophenoxyacetate alpha-ketoglutarate dioxygenase activity(GO:0018602) hypophosphite dioxygenase activity(GO:0034792) gibberellin 2-beta-dioxygenase activity(GO:0045543) C-19 gibberellin 2-beta-dioxygenase activity(GO:0052634) C-20 gibberellin 2-beta-dioxygenase activity(GO:0052635) |
| 0.1 | 4.4 | GO:0052771 | coenzyme F390-A hydrolase activity(GO:0052770) coenzyme F390-G hydrolase activity(GO:0052771) |
| 0.1 | 0.7 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 20.4 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 4.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 1.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 11.7 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.1 | 2.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 1.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 6.4 | GO:0008186 | ATP-dependent RNA helicase activity(GO:0004004) RNA-dependent ATPase activity(GO:0008186) |
| 0.1 | 5.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 17.3 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 2.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 4.6 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.6 | GO:0004889 | acetylcholine-activated cation-selective channel activity(GO:0004889) |
| 0.1 | 0.6 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 2.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.1 | 14.2 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 17.3 | GO:0001077 | transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) |
| 0.1 | 5.6 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 22.9 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 2.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 20.0 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 4.1 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 3.3 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 2.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 1.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) |
| 0.0 | 13.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 4.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.3 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.9 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.4 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 3.0 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 1.3 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |